Figure 1.
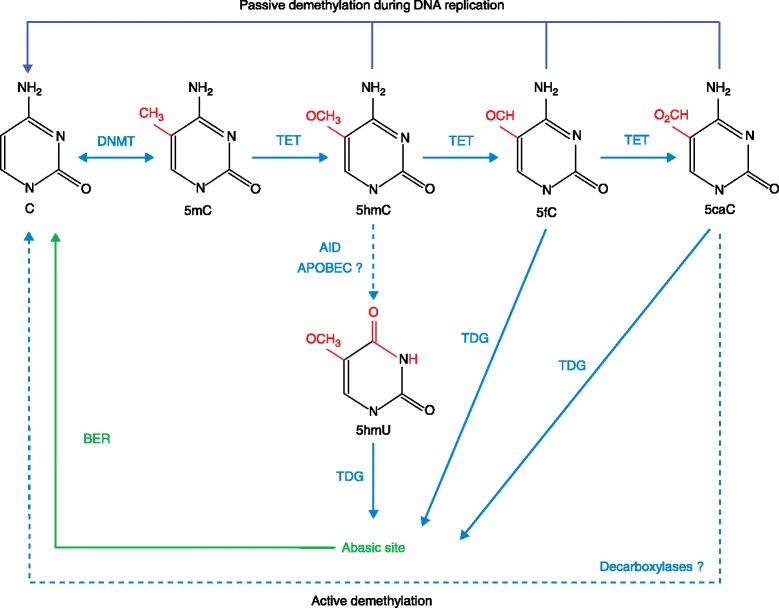
Regulation of DNA methylation and demethylation. DNA demethylation can occur spontaneously via the DNMT enzymes that methylated the nucleotide cytosine (5-methylcytosine, 5-mC) originally. A passive replication-dependent mechanism of DNA methylation is also possible. Several active demethylation pathways have been postulated. TET family proteins catalyze the oxidation of 5-mC into 5-hydroxymethylcytosine (5-hmC) and can further oxidize 5-hmC to 5-formylcytosine (5-fC) and 5-carboxycytosine (5-caC). 5-hmC recognition and transformation into 5-hydroxymethyluracyl (5-hmU) by activation-induced deaminase (AID) to facilitate repair by DNA glycosylase and the base-excision repair (BER) pathway is still controversial. These latter activities are also thought to process 5-fC and 5-caC into unmodified cytosine . The decarboxylases involved in this process are still to be identified. APOBEC, apolipoprotein B mRNA editing enzyme; DNMT, DNA methyltransferase; T, thymine; TDG, thymine DNA glycosylase; TET, ten-eleven translocation.
