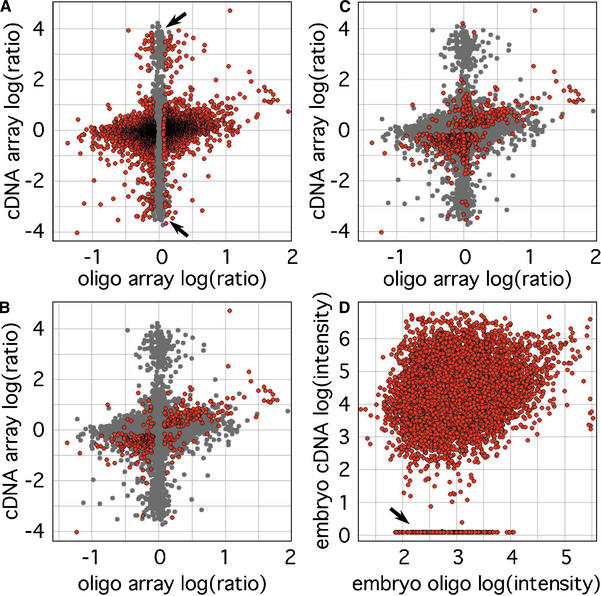
Figure 2.
Scatter plots comparing log expression ratios in mouse E12.5 embryo and placenta measured by 60-mer oligo and cDNA microarrays. Each marker represents averaged results from dye-swapped duplicate microarrays using three biological replicates. (A) Probes showing significant differential expression by ANOVA-FDR analysis of oligo microarray measurements are indicated in red (n = 5200, FDR < 0.05, correlation coefficient [cc] = 0.27, slope = 0.30). (B) Significant transcripts identified by cDNA array show better agreement (n = 545, P < 0.05, cc = 0.52, slope = 0.32), and (C) the intersection of these two sets are even more closely matched (n = 336, cc = 0.67, slope = 0.51). (D) A number of transcripts were measured at background intensity by cDNA arrays, but had substantial intensities on oligo microarrays, as indicated by the arrow. This group was the major component of two sets of log(ratio) outliers, indicated by the arrows in A.