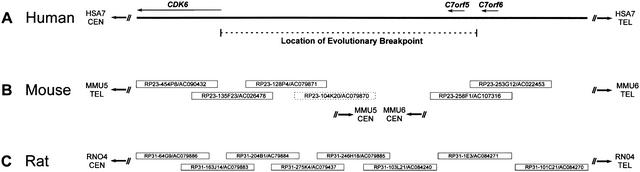
Figure 1.
Physical mapping of the human, mouse, and rat genomic regions flanking an evolutionary breakpoint. The depicted segment of HSA7q21 (A) and the orthologous region of RNO4 (C) are contiguous in each species; however, the orthologous segment in the mouse genome is split between two regions (B), with portions located at the proximal (i.e., centromeric) ends of MMU5 and MMU6. The indicated location of the evolutionary breakpoint on HSA7q21 is based on genetic and cytogenetic mapping studies (Thomas et al. 1999). The positions of the three genes (CDK6, C7orf5, and C7orf6) on HSA7q21 are based on finished genomic sequence (GenBank nos. AC000065, AC004128, AC004011, AC002454, and AC000119). Also shown are the minimal tiling paths of BACs selected for sequencing the orthologous regions on MMU5, MMU6, and RNO4 (Thomas et al. 1999; Summers et al. 2001), with the clone name/GenBank accession number indicated for each. Note that the mouse BACs depicted here are the most proximal clones in larger (20-BAC) tiling paths assembled for both MMU5 and MMU6 (data not shown). One clone (RP23–104K20) was subsequently found not to map to MMU5 (indicated by a dashed box). The orientation of the depicted sequences and clones relative to each telomere (TEL) and centromere (CEN) is indicated. Note that the human sequence as well as the mouse and rat clones are not drawn to scale.