Table 2.
Comparison of Genome Coverage
| chr20 | CDS | 3′UTR | 5′UTR | upstream | |
| Blastz all | 40.5% | 98.5% | 87.1% | 89.0% | 87.2% |
| Blastz tight | 5.6% | 92.5% | 26.0% | 39.6% | 28.3% |
| PH all | 29.7% | 95.5% | 55.0% | 59.3% | 52.5% |
| PH tight | 5.0% | 91.2% | 25.1% | 36.3% | 25.2% |
| transl. BLAT | 5.8% | 90.3% | 29.2% | 38.4% | 27.2% |
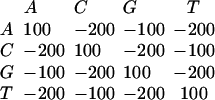 |
a gap open score of −2000, a gap extension score of −50, and a threshold of 3400.
