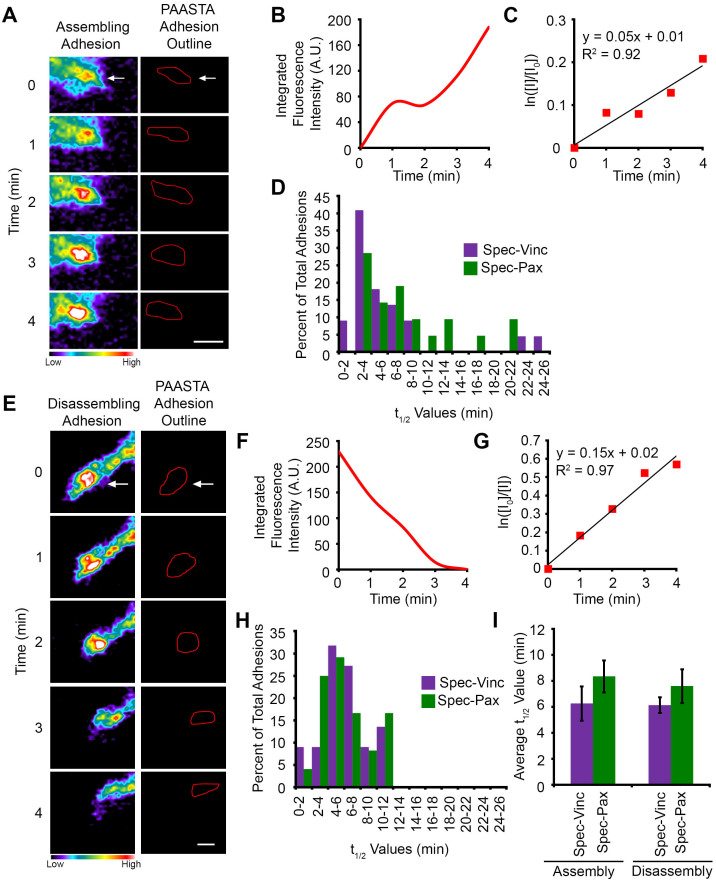
Figure 6. Analysis of adhesion dynamics for U2OS cells embedded in 3D type I collagen matrices using PAASTA.
(A,E) Left, Time-lapse images of an assembling (panel A) and a disassembling (panel E) adhesion (white arrows) in Spec-Pax expressing cells embedded in a 3D type I collagen matrix are shown. The images are shown in pseudo-color coding to indicate the range of fluorescence intensities. Warm colors represent higher fluorescence intensities while cool colors denote lower fluorescence intensities. Right, Outlines of the adhesions as segmented by PAASTA. (B,F) Graphs of the fluorescence intensities of the assembling (panel B) and disassembling (panel F) adhesion are shown for PAASTA adhesion tracking. (C,G) Plots of the natural log of the fluorescence intensity of the adhesions at given time points (I) relative to the initial fluorescence intensity (I0) are shown. Trendlines with the corresponding equation (y = mx + b) and R2 values are shown. The slopes of these graphs (m) are the apparent rate constant for adhesion assembly (panel C) or the rate constant for adhesion disassembly (panel G). (D,H) Histograms of the distribution of t1/2 values for adhesion assembly (panel D) and disassembly (panel H) are shown for cells expressing either Spec-Pax or Spec-Vinc. (I) The average t1/2 values for adhesion assembly and disassembly are shown for cells expressing either Spec-Pax or Spec-Vinc. S.E.M. was calculated from 47 adhesions (21 assembly, 26 disassembly) for Spec-Pax and 44 adhesions (22 assembly, 22 disassembly) for Spec-Vinc. A total of 9 cells were used for the analysis of adhesion assembly and disassembly. A Wilcoxon rank sum test showed no statistically significant difference between Spec-Pax and Spec-Vinc for assembly (Z = −1.269, p = 0.205) or disassembly (Z = −0.406, p = 0.685). For panels A and E, Bar, 1 μm.