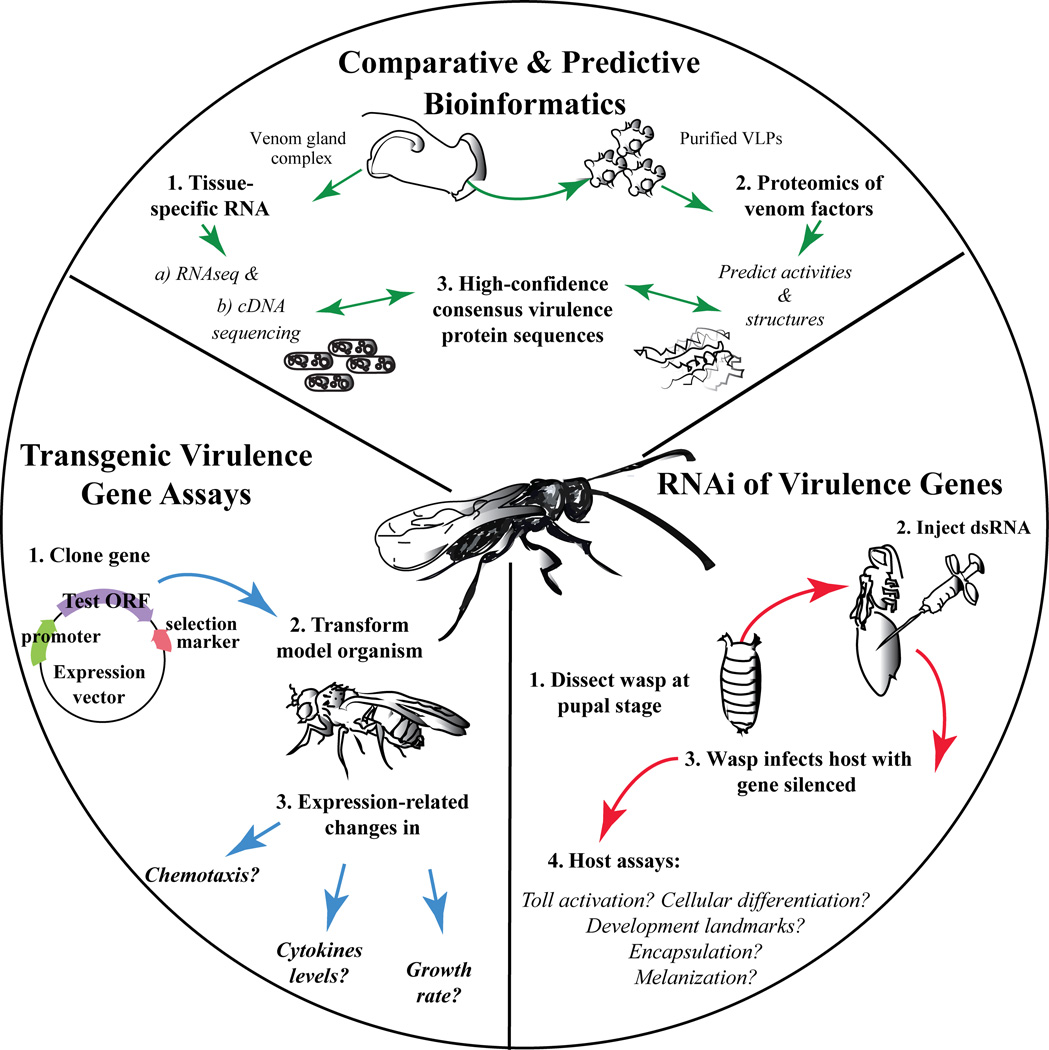
Figure 2. Structural and functional parasitoid venomics.
(Top sector) Comparative and predictive bioinformatics: Either total RNA or mRNA from venom glands is sequenced. In addition, proteomes of purified immune-suppressive factors (e.g., from venom fluid or purified VLPs) are matched with RNA and genomic sequences, if available. High-confidence matches and consensus sequences are fed into structural bioinformatics algorithms. Structural folds, domain architectures, catalytic motifs, etc., are predicted to provide functional insight, especially for novel and un-annotated proteins. (Right bottom sector) RNAi of virulence genes: For high- or medium-throughput analysis of virulence proteins of interest, newly demonstrated RNAi approaches would be appropriate. Blocking translation of single virulence proteins can provide specific biological activity information given non-redundant functions in hosts. (Left bottom sector) Transgenic virulence gene assays: For proteins with clear-cut functions in blocking host immunity, spatially and temporally controlled in vivo transgenic virulence, development, and other assays can be used in Drosophila and in other model systems to assess molecular and signaling targets.