Figure 3. Increased volatility in microbiota of T5KO mice.
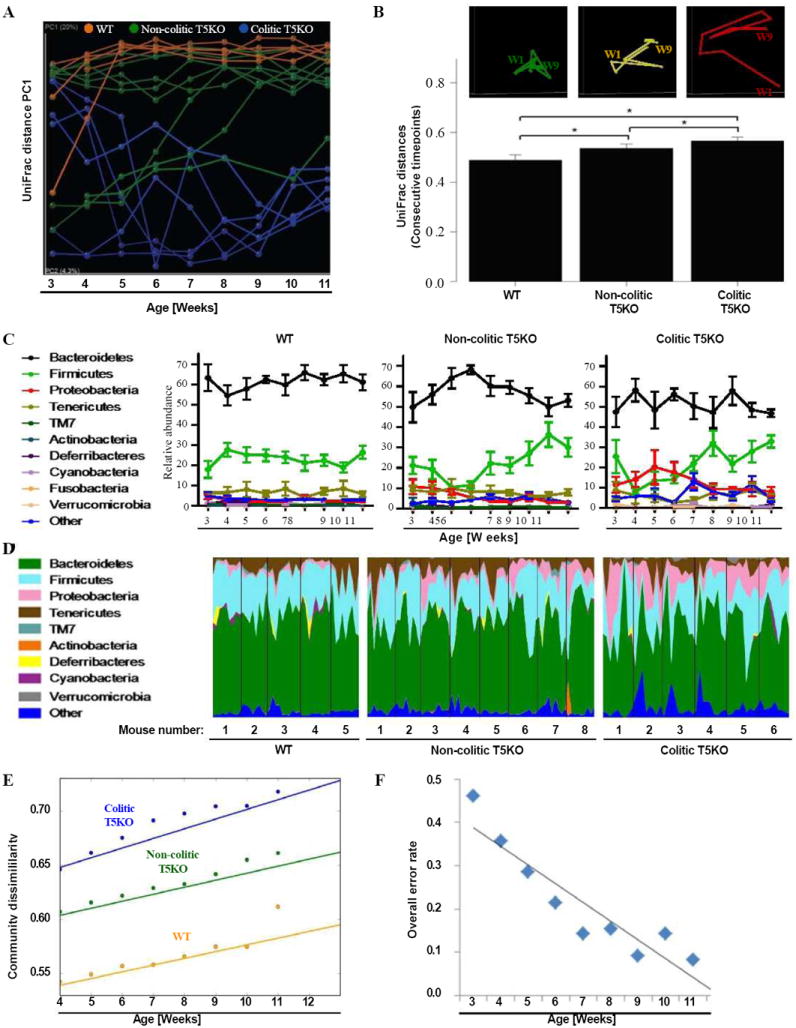
Stool from wild-type, non-colitic and colitic T5KO mice (n=5-8 mice per group) were collected weekly for 9 weeks after weaning (from 3-week to 11-week old). Stool microbiota composition was analyzed via 16S rRNA analysis. (A) Mouse cecal bacterial communities were clustered using principal coordinates analysis (PCoA) of the UniFrac unweighted distance matrix. PC1 is plotted for each time point (from 3-week to 11-week old). The time is expressed on the x-axis and the percentage of the variation explained by the plotted principal coordinates is indicated in the y-axis labels. Results are from an analysis of 5-8 mice per group (samples generating less than 1000 sequences were removed from analysis). (B) After clustering of mouse cecal bacterial communities using principal coordinates analysis (PCoA) of the UniFrac unweighted distance matrix, a representative mouse has been used to illustrate the time point evolution of the microbiota (top panel). The average of the UniFrac unweighted distance for each category (WT, non-colitic and colitic T5KO) between consecutive time points has been calculated (bottom panel). (C) Relative abundance of phyla in stool bacteria from WT (left panel), non-colitic T5KO (middle panel) and colitic T5KO (right panel) mouse group. (D) Relative abundance of phyla over time (from 3-week to 11-week old) in stool bacteria from individual WT (left panel), non-colitic T5KO (middle panel) and colitic T5KO (right panel) mice. (E) Semivariogram plot of community dissimilarity (UniFrac, y axis) vs. days dissimilarity (Euclidean, x axis). (F) Overall misclassification error rates using 9 families at each week. Analysis was done by ANOVA and statistical significance (P<0.01) is denoted by asterisk (*). Related materials can be found in supplementary figure 1.
