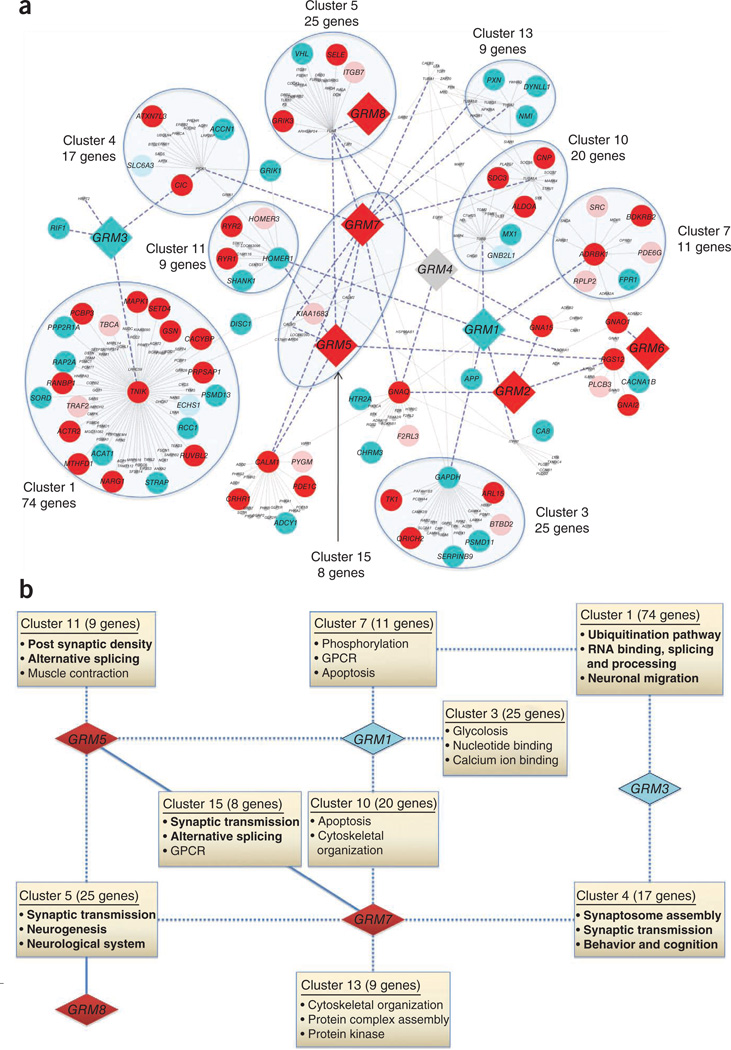
Figure 2.
GRM receptor gene interaction networks affected in ADHD. (a) GRM receptor genes are shown as large diamond-shaped nodes, and other genes within two degrees of interaction with GRM genes are shown as smaller circular nodes. Nodes are colored to represent the enrichment of the CNVs: dark red represents deletions enriched in cases, light red represents deletions enriched in controls, dark turquoise represents duplications enriched in cases, light turquoise represents duplications enriched in controls, and gray represents diploids that are devoid of CNVs. Thick blue dashed lines highlight edges that are connected to at least one GRM gene, and thin gray lines represent all other gene interactions. Highly connected modules enriched for significant functional annotations are highlighted by blue shaded ellipses. Details on the gene-based CNV observations are included in Supplementary Table 16, and the respective gene functional clusters are listed in Supplementary Table 17. (b) A schematic overview showing the interaction of GRM receptors affected in ADHD with modules of genes enriched for functional significance. GRM receptor genes are shown as diamonds colored either turquoise or red to represent duplications and deletions, respectively, that were enriched in cases. Boxes highlight the functional modules defined by the network of interacting genes (a) that are significantly enriched for Gene Ontology annotations. The functional modules describe significant functional annotations and are labeled with the cluster name and the number of component genes in parenthesis. Functional annotations that may be particularly pertinent to the underlying pathophysiology of ADHD are shown in bold. The edges of the network connect GRM receptor genes to functional modules: solid lines indicate membership of the GRM-interacting gene in the functional module, and dotted lines indicate a first-degree relationship between GRM receptor genes and at least one component gene of a functional module