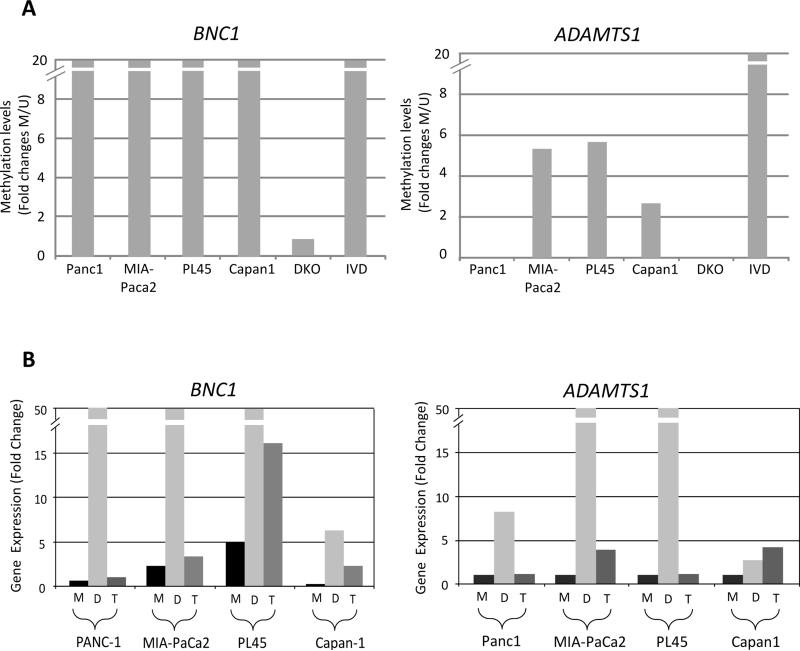
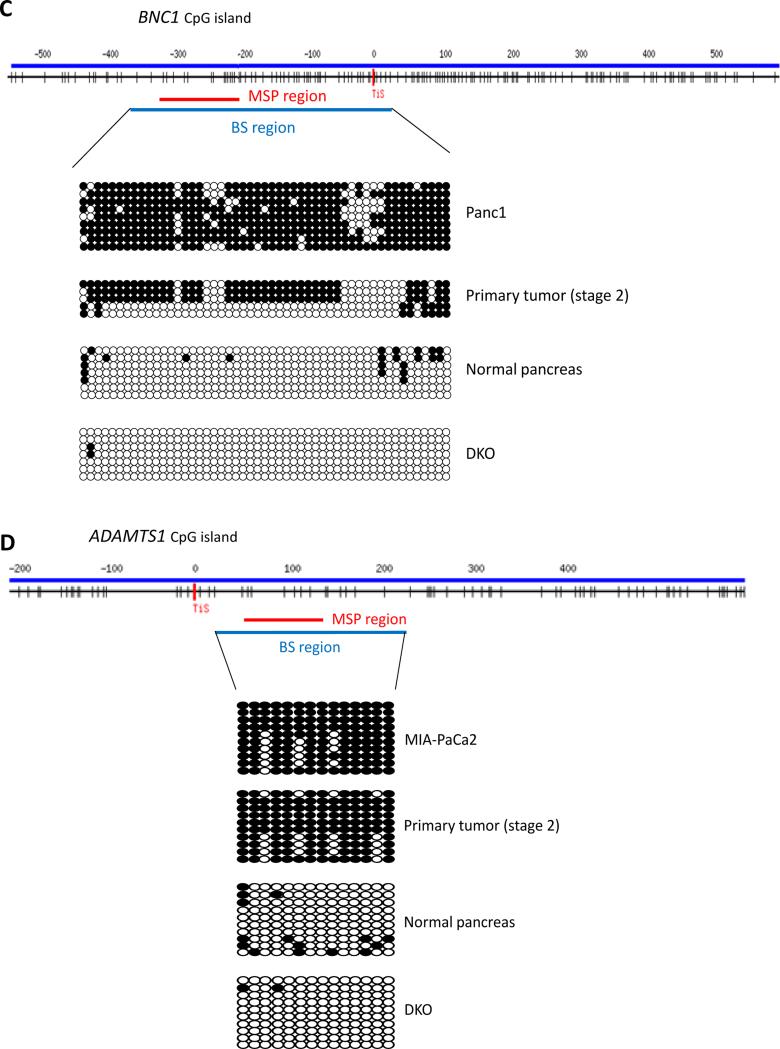
Fig. 2.
(A-B) Silencing of BNC1 and ADAMTS1 genes in pancreatic cancer cell lines. (A) qMSP analysis of BNC1 and ADAMTS1 gene promoter region and correlation with (B) gene expression by qPCR in pancreatic cancer cell lines. IVD= in vitro methylated DNA. DKO=double knockout (DKO) HCT116 cells (DNMT1−/− and DNMT3b−/−). Quantitative PCR expression is shown as fold change ± standard error relative to mock-treated (M) cells during 5 μM 5-Aza-2’-deoxycytidine (DAC; D) and 300 nM Trichostatin A (TSA; T) treatments. Normal pancreas and DKO cells were used as controls. (C-D) Bisulfite sequencing analysis of CpG island in (C) BNC1 and (D) ADAMTS1 gene promoter region. Bisulfite sequencing analysis of the BNC1 (for Panc1) and ADAMTS1 (for MIA-PaCa2) genes in a pancreatic cancer cell line, a primary pancreatic cancer (stage II), normal pancreatic tissue and DKO cell line as a negative control. Open circles represent unmethylated CpG sites and filled circles represent methylated CpG sites with each row representing a single clone. TIS indicate transcriptional start site. Location of CpG sites (BST: upstream region from −331 to +36 for BNC1, +22 to +207 for ADAMTS1 relative to transcriptional start site). The red and blue bars represent MSP amplification and bisulfite amplification region, respectively.