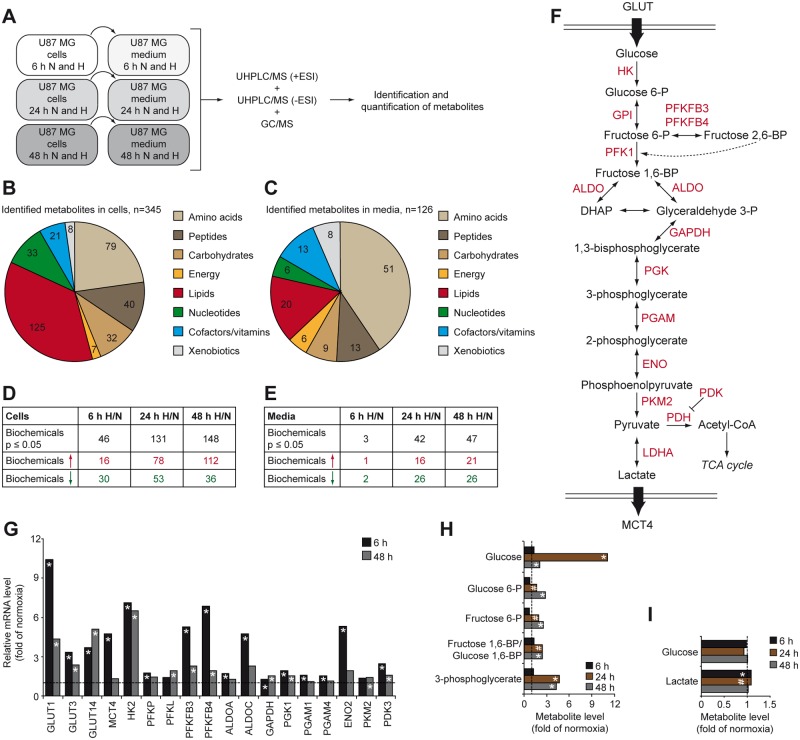
Fig 1. The metabolic phenotype of hypoxic glioblastoma cells.
A, U87 MG cells were grown at normoxic (N, 21% O2) or hypoxic (H, 1% O2) conditions for 6, 24 or 48 h. Cells (n = 6 for each time point) and the corresponding conditioned media (n = 6 for each time point) were analyzed on three separate mass spectrometry platforms. The identification of known chemical entities was based on comparison to metabolic library entries of purified standards. B and C, Class distribution of identified metabolites in cells (B) and conditioned media (C). D and E, Summary of number of biochemicals that was significantly down- or up-regulated in hypoxic cells (D) and corresponding conditioned media (E) when compared with normoxic conditions. F, Schematic diagram of glycolysis. G, Hypoxia-driven transcriptional activation of genes encoding proteins involved in glycolysis at the indicated time-points. Data represent fold change of mRNA levels in hypoxic vs. normoxic cells. H and I, Hypoxic modulation of glycolysis associated metabolites. Data represent fold change of metabolite levels in hypoxic GBM cells (H) and their conditioned media (I) compared with normoxic samples. * P < 0.05; # 0.05 < P < 0.1. ALDO, aldolase; ENO2, enolase 2; GPI, glucose-6-phosphate isomerase; HK, hexokinase; LDHA, lactate dehydrogenase A; PDK3, pyruvate dehydrogenase kinase 3; PFK, phosphofructokinase; PFKFB, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase; PGAM, phosphoglycerate mutase; PGK1, phosphoglycerate kinase 1; PKM2, pyruvate kinase isoenzyme type-M2.