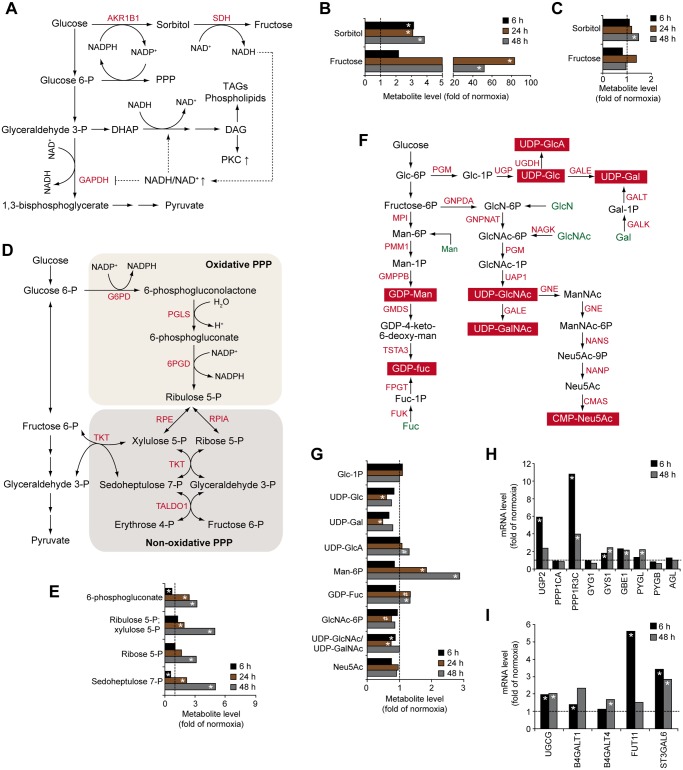
Fig 2. Altered glucose shunting in hypoxic GBM cells.
A, Schematic of the polyol pathway. B and C, Hypoxic effects on metabolites of the polyol pathway in cells (B) and media (C). D, Schematic of the pentose phosphate pathway (PPP). E, Hypoxic modulation of PPP metabolites in cells. F, Schematic of metabolic pathways involved in nucleotide sugar synthesis. G, Hypoxic modulation of nucleotide sugars and metabolites involved in their synthesis. H and I, Hypoxic induction of mRNAs encoding enzymes involved in glycogen synthesis (H) and protein and lipid glycosylation (I) at the indicated time-points. Data represent fold change in hypoxic compared with normoxic cells. * P < 0.05; # 0.05 < P < 0.1. CMAS, cytidine monophosphate N-acetylneuraminic acid synthetase; FPGT, fucose-1-phosphate guanylyltransferase; FUK, fucose kinase; GALE, UDP-galactose-4 epimerase; GALK, galactokinase; GALT, galactose-1-phosphate-uridylyltransferase; GMDS, GDP-mannose 4,6-dehydratase; GMPPB, GDP-mannose pyrophosphorylase B; GNE, glucosamine (UDP-N-Acetyl)-2-epimerase/N-acetylmannosamine kinase; GNPDA, glucosamine-6-phosphate isomerase; GNPNAT, glucosamine-phosphate N-acetyltransferase 1; G6PD, glucose-6-phosphate dehydrogenase; MPI, mannose phosphate isomerase; NAGK, N-acetylglucosamine kinase; NANP, N-acetylneuraminic acid phosphatase; NANS, N-acetylneuraminic acid synthase; 6PGD, phosphogluconate dehydrogenase; PGLS, 6-phosphogluconolactonase; PGM, phosphoglucomutase; PMM1, phosphomannomutase 1; RPE, ribulose-phosphate 3-epimerase; RPIA, ribose-5-phosphate isomerase; TALDO1, transaldolase 1; TKT, transketolase; TSTA3, GDP-L-fucose synthetase; UAP1, UDP-N-acteylglucosamine pyrophosphorylase 1; UGDH, UDP-glucose 6-dehydrogenase; UGP, UDP—lucose pyrophosphorylase.