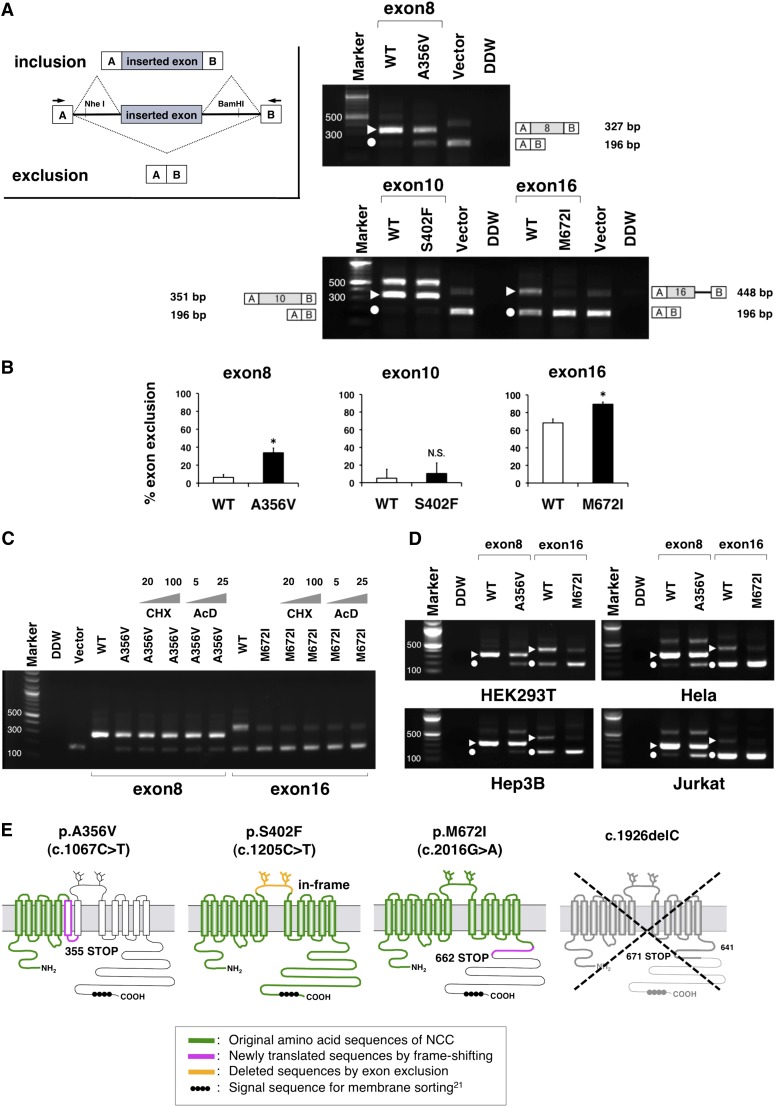
Figure 2.
Hybrid minigene assay reveled the aberrant exonic mRNA splicing in GS syndrome. (A) RT-PCR amplified products of hybrid minigene transcripts in HEK293T cells. The transcripts produced by the hybrid minigene are schematically shown and the arrows show the primers used to amplify (inset). The triangles and circles indicate the transcripts of exon inclusion and the exclusion, respectively. The sizes of the exon-included transcripts included 327 nucleotides for exon 8, 351 for exon 10, and 448 for exon 16. There were some defectively spliced fragments including partial introns on the exon 10 lane (575 nucleotides) and on the vector lane (407 nucleotides). The identity of each fragment is illustrated schematically on the side. (B) Quantification of the splicing percentage in the graph was densitometrically calculated on a molar basis as the percentage of exclusion (%)=(lower band/[lower band+upper band])×100.15 Error bars represent SEM (n=3). *P<0.05, unpaired t test. (C) The treatment with de novo protein synthesis inhibitors indicates that the transcriptional products are stable. The gray triangles represent the concentrations of the reagents in a dose-dependent manner. AcD, actinomycin D; CHx, cycloheximide. (D) Comparison among the other cell types in the RT-PCR products of the minigene transcripts. Transfection of the hybrid minigenes to Hela, Hep3B, and Jurkat cell lines was performed the same as for HEK293T. The triangles and circles indicate the transcripts of exon inclusion and exclusion, respectively. (E) Predicted protein structures translated from the aberrant transcripts of the exon exclusions. Once the c.1067C>T mutation induces the exon8 exclusion, the frameshift and prematurely terminated codon occur in the transmembrane domain. The c.2016G>A mutation also induces the exon16 exclusion, resulting in a truncated protein that lacks a large part of the carboxyl terminal domain, which contains the signal sequence required for normal translocation. The c.1926delC allele is expected to be unstable because of mRNA degradation.