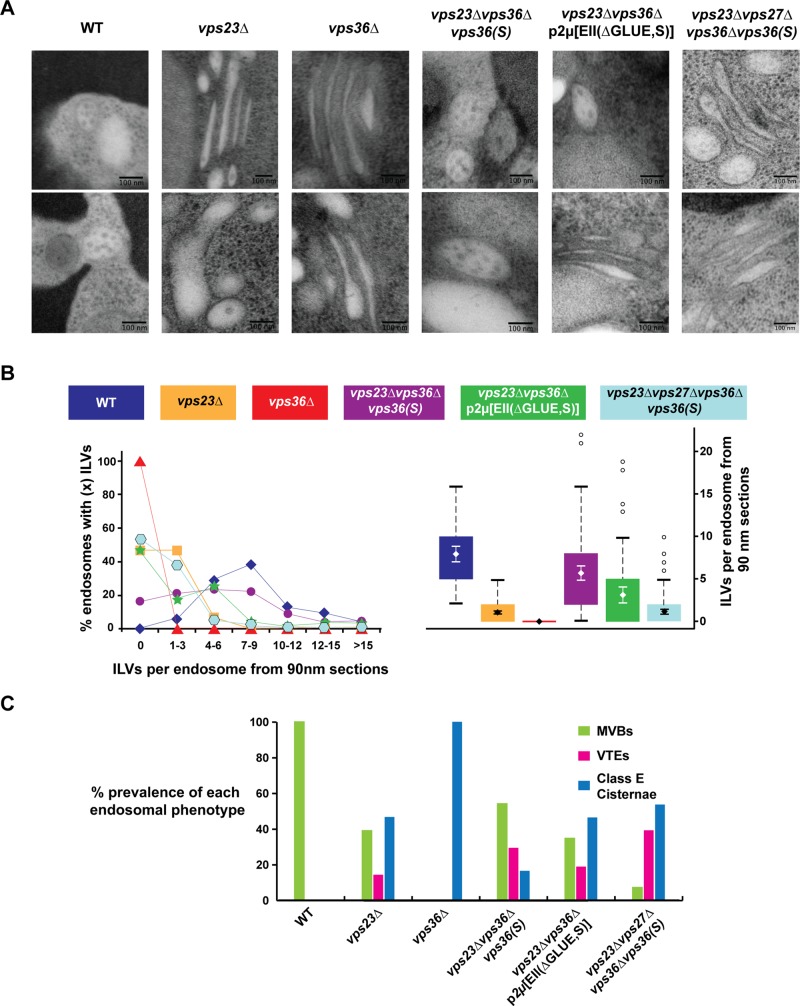
FIGURE 4:
TEM analysis of MVB formation. The strains vps23∆vps36∆vps36(S), vps23∆vps36∆p2μ[EII(∆GLUE,S)], and vps23∆vps27∆vps36∆vps36(S) lack the genomic VPS36 but instead express WT or mutant copy of VPS36/ESCRT-II under its native promoter from CEN/2μ plasmids. (A) Representative images of endosomes for each strain. (B) ILVs per endosome from 90-nm thin sections shown as a distribution and box plot. The error bars show the 95% confidence interval over the mean. All differences, except between the strains vps23∆ and vps23∆vps27∆vps36∆vps36(S), are significant (with p < 0.001). (C) Endosomal morphology distribution. Number of endosomal profiles counted for each strain: 52, 105, 34, 109, 69, and 97 (in the order of strain presentation).