Changing light and copper conditions jointly regulate the expression of a large cohort of genes; the microRNA MIR408 plays a pivotal role in this regulation.
Abstract
Light and copper are important environmental determinants of plant growth and development. Despite the wealth of knowledge on both light and copper signaling, the molecular mechanisms that integrate the two pathways remain poorly understood. Here, we use Arabidopsis thaliana to demonstrate an interaction between SQUAMOSA PROMOTER BINDING PROTEIN-LIKE7 (SPL7) and ELONGATED HYPOCOTYL5 (HY5), which mediate copper and light signaling, respectively. Through whole-genome chromatin immunoprecipitation and RNA sequencing analyses, we elucidated the SPL7 regulon and compared it with that of HY5. We found that the two transcription factors coregulate many genes, including those involved in anthocyanin accumulation and photosynthesis. Moreover, SPL7 and HY5 act coordinately to transcriptionally regulate MIR408, which results in differential expression of microRNA408 (miR408) and its target genes in response to changing light and copper conditions. We demonstrate that this regulation is tied to copper allocation to the chloroplast and plastocyanin levels. Finally, we found that constitutively activated miR408 rescues the distinct developmental defects of the hy5, spl7, and hy5 spl7 mutants. These findings revealed the existence of crosstalk between light and copper, mediated by a HY5-SPL7 network. Furthermore, integration of transcriptional and posttranscriptional regulation is critical for governing proper metabolism and development in response to combined copper and light signaling.
INTRODUCTION
Essentially sessile in nature, plants have evolved sophisticated mechanisms to maintain metabolic homeostasis and a remarkable capacity to reprogram development when resource levels vary. Light and copper are among the most important environmental factors for plant growth. In addition to providing a source of energy, light regulates many plant processes and induces massive reprogramming of the transcriptome (Chen et al., 2004; Jiao et al., 2007). Genetic and molecular approaches in Arabidopsis thaliana have identified many regulatory factors and pathways required for proper light signaling. ELONGATED HYPOCOTYL5 (HY5) encodes a bZIP-type transcription factor that functions downstream of multiple photoreceptors to promote photomorphogenesis (Oyama et al., 1997; Ang et al., 1998). Detailed characterization of the hy5 mutants revealed myriad phenotypic defects, including elongated hypocotyl, reduced pigment content, aberrant chloroplast development, altered root morphology, and compromised hormonal responses (Oyama et al., 1997; Cluis et al., 2004; Vandenbussche et al., 2007). HY5 binds to G-box-like motifs in the light-responsive promoters (Chattopadhyay et al., 1998; Yadav et al., 2002; Shin et al., 2007; Song et al., 2008). Chromatin immunoprecipitation (ChIP) coupled tiling microarray analysis showed that HY5 binds to ∼40% of the coding loci in the Arabidopsis genome and detectably impacts the expression level of ∼3000 genes (Lee et al., 2007; Zhang et al., 2011).
As a transition metal, copper is an essential cofactor for numerous proteins. The most abundant copper protein in plants is plastocyanin (PC), which transfers electrons from the cytochrome b6f complex to photosystem I (Burkhead et al., 2009). Copper is also used as a cofactor by plant proteins involved in neutralizing reactive oxygen species, lignification of the cell wall, ethylene perception, and formation of phenolics in response to pathogens (Burkhead et al., 2009). Regulating the abundance of these proteins is important to maintain copper homeostasis and prioritize the use of cellular copper in plants. Studies in the green alga Chlamydomonas reinhardtii revealed COPPER RESPONSE REGULATOR1 (CRR1) as a zinc finger transcription factor that is specifically activated under copper deficiency (Kropat et al., 2005). The GTAC motif found in CRR1 targets is recognized as the core of copper-response elements in diverse plants (Quinn and Merchant, 1995; Quinn et al., 1999; Kropat et al., 2005; Nagae et al., 2008; Yamasaki et al., 2009). In Arabidopsis, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE7 (SPL7) is orthologous to CRR1 (Yamasaki et al., 2009). In spl7 mutants, many of the genes related to copper homeostasis, including the transporters COPT1 and COPT2, the copper chaperones CCH and CCS, as well as the copper/zinc superoxide dismutases CSD1 and CSD2, are dysregulated (Yamasaki et al., 2009; Bernal et al., 2012). Copper specifically inhibits the DNA binding activity of both CRR1 and SPL7 and prevents transcription activation in vitro (Sommer et al., 2010). Thus, Arabidopsis SPL7 likely functions as a copper sensor that regulates gene expression in response to changing cellular copper levels (Sommer et al., 2010).
In addition to transcriptional regulators, microRNAs (miRNAs) are emerging as a class of sequence-specific, trans-acting regulatory small RNA molecules that modulate gene expression mainly at the posttranscriptional level. After processing from stem-loop-structured precursors and integration into the RNA-induced silencing complex, miRNAs function in general as gene repressors by directing the cleavage or translational repression of the target transcripts (Voinnet, 2009). The large numbers of miRNAs in eukaryotes entail an increased complexity of gene regulatory mechanisms. In plants, miRNAs have been implicated in light and copper signaling. A global ChIP analysis also observed HY5 occupancy at the promoter region of eight miRNAs (Zhang et al., 2011). Furthermore, several miRNAs were predicted to target transcripts encoding copper proteins (Jones-Rhoades and Bartel, 2004; Sunkar et al., 2006; Yamasaki et al., 2007; Abdel-Ghany and Pilon, 2008). Their expression was induced by copper deficiency in an SPL7-dependent manner (Yamasaki et al., 2009; Bernal et al., 2012). It was thus hypothesized that these miRNAs repress nonessential copper proteins (such as CSD1 and CSD2) and their chaperone to save copper for essential proteins such as PC under impending deficiency (Yamasaki et al., 2007).
However, how light and copper signals work together to regulate gene expression for optimal plant growth and development is poorly understood. In this work, we demonstrate the interaction between SPL7 and HY5, providing evidence for the integration of copper and light signaling. By elucidating the SPL7 regulon through ChIP and RNA sequencing and comparing it with the previously identified HY5 regulon (Zhang et al., 2011), we found that these two transcription factors have significant overlap in terms of the genes they bind directly and the genes whose expression levels they regulate. Furthermore, we show that microRNA408 (miR408) is a critical component of the SPL7-HY5 network and that constitutively activated miR408 rescues the developmental defects of the hy5, spl7, and hy5 spl7 mutants. These findings thus revealed a molecular basis incorporating transcriptional and posttranscriptional regulations for coordinated plant responses to changing copper and light regimens.
RESULTS
SPL7 Interacts with HY5
We observed that Arabidopsis seedlings respond decisively to combined light and copper regimes when grown under relatively high light (HL; 170 µmol m−2 s−1) or low light (LL; 40 µmol m−2 s−1) intensity and sufficient copper (SC; 5 μM) or deficient copper (DC; 0.1 μM) conditions (Supplemental Figure 1). Plant morphology was primarily influenced by light, with seedlings grown in HL having shortened hypocotyls and roots but increased fresh weight (Supplemental Figures 2A and 2B). However, both light and copper influenced the contents of essential metabolites such as chlorophyll, anthocyanin, and glucose (Supplemental Figures 2C to 2E). Overall, seedlings displayed distinctive morphological and metabolic profiles under the four combinations of light and copper conditions (Supplemental Figure 2). These results indicate that there is crosstalk between the light and copper signaling pathways for orchestrating plant growth and metabolism.
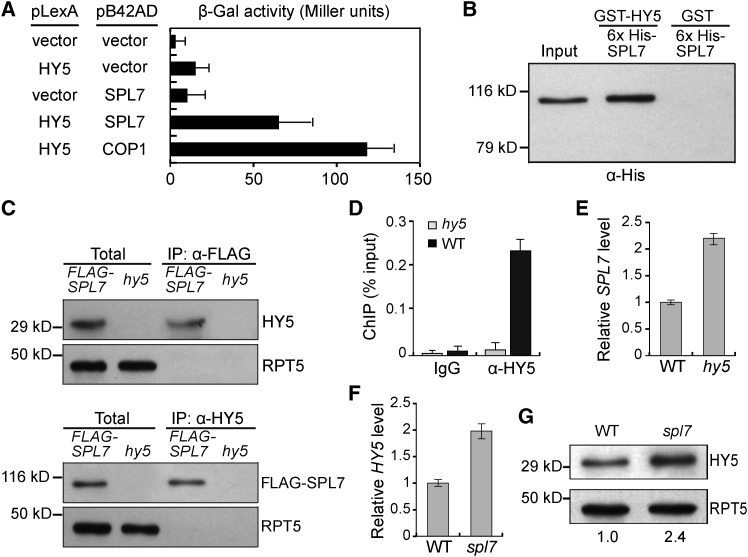
To begin elucidating the light-copper crosstalk, we examined the developmental expression profiles of HY5 and SPL7 in Arabidopsis and found that both are expressed in the six examined organ types (Supplemental Figure 3A). This result prompted us to investigate whether the two transcription factors interact with each other. We performed a yeast two-hybrid assay in which HY5 and SPL7 were fused in-frame to the binding domain of LexA and the activation domain of B42, respectively. Specifically, increased reporter activity was observed when both the HY5 and SPL7 fusion proteins were expressed in the same yeast cells (Figure 1A). We also performed an in vitro pull-down assay using recombinant SPL7 and HY5, which showed that GST-tagged HY5, but not GST alone, was able to pull down 6×His-tagged SPL7 (Figure 1B). Furthermore, we generated transgenic Arabidopsis plants expressing N-terminal FLAG-tagged SPL7 driven by the cauliflower mosaic virus 35S promoter (35S:FLAG-SPL7). Using these plants, we performed a coimmunoprecipitation assay and found that the anti-FLAG and the anti-HY5 antibodies could specifically pull down HY5 and FLAG-SPL7 from plant extracts, respectively (Figure 1C). Together, these results demonstrate the SPL7-HY5 physical interaction.
Figure 1.
Interaction of SPL7 and HY5.
(A) HY5 interacts with SPL7 in a yeast two-hybrid assay. The β-galactosidase activities resulted from the HY5-SPL7 interaction, and various controls are shown. The HY5-COP1 interaction (Ang et al., 1998) was used as a positive control. Error bars indicate sd (n = 4).
(B) HY5 can pull down SPL7 in vitro. Purified 6×His-tagged SPL7 was incubated with recombinant GST-HY5 or GST and immunoprecipitated with agarose beads conjugated with a GST antibody. The precipitates were subjected to immunoblotting using the anti-His antibody. Input, 5% of the purified SPL7 used in the pull-down assays.
(C) HY5 associates with SPL7 in vivo. Total protein extracts from 35S:FLAG-SPL7/spl7 seedlings were incubated with anti-FLAG or anti-HY5 antibody-conjugated agarose beads. The precipitates and total extracts were subjected to immunoblotting with antibodies against HY5 and FLAG, respectively. RPT5 was used as a control.
(D) Confirmation of HY5 binding to the SPL7 promoter by ChIP-qPCR. ChIP was performed in wild-type and hy5 seedlings with or without the anti-HY5 antibody. The resultant DNA was analyzed by quantitative PCR with the values normalized to their respective DNA inputs.
(E) and (F) RT-qPCR analyses of SPL7 (E) and HY5 (F) transcript levels. Reverse-transcribed cDNA from wild-type, hy5, or spl7 seedlings was examined by quantitative PCR with the values normalized to those of the wild type.
(G) Immunoblot analysis of HY5 protein levels in wild-type and spl7 seedlings. Values below the blots represent HY5 levels normalized against the loading control RPT5 using ImageJ and set to 1 for the wild type.
Next, we tested whether SPL7 and HY5 influence each other’s expression. We extracted the 2 kb of sequence upstream of the transcription start site (TSS) of SPL7 and HY5 as an approximation of their promoters. Scanning the SPL7 promoter identified a cluster of five HY5 binding motifs, which coincides with a strong HY5 binding peak at this region according to the whole-genome occupancy data (Zhang et al., 2011; Supplemental Figure 3B). This observation was confirmed by electrophoretic mobility shift assay (EMSA) using recombinant HY5 and a DNA fragment of the SPL7 promoter containing all five motifs as the probe (Supplemental Figure 3C) as well as quantitative PCR analysis of the SPL7 promoter region following ChIP (ChIP-qPCR) by the anti-HY5 antibody (Figure 1D). Monitoring transcript levels by reverse transcription coupled with quantitative PCR (RT-qPCR) revealed that SPL7 expression increases over 2-fold in hy5 mutants (Figure 1E). These results indicate that HY5 directly binds to the SPL7 promoter and negatively regulates its transcription.
Scanning the promoter region of HY5 revealed no GTAC motifs. Consistent with this, ChIP-qPCR analysis revealed that SPL7 does not bind to the HY5 promoter. However, we found that HY5 abundance showed an over 2-fold increase in the spl7 mutant compared with the wild type at both the mRNA and protein levels (Figures 1F and 1G), indicating that HY5 is negatively regulated by SPL7. Together, these results demonstrate that SPL7 and HY5 interact with each other both physically and genetically, which suggests a feedback mechanism for linking the light- and copper-responsive gene networks.
Genome-Wide Analyses of the SPL7 Regulon
To facilitate the global identification of SPL7 binding sites in Arabidopsis, we generated transgenic lines expressing FLAG-tagged SPL7 in the spl7 mutant background (35S:FLAG-SPL7/spl7). Characterization of the transgenic lines indicated that FLAG-SPL7 is properly expressed, levels of SPL7-regulated miR398 (Yamasaki et al., 2009) and miR408 (Zhang and Li, 2013) are restored, and growth defects of spl7 (reduced fresh weight and shorter root when grown under DC) are rescued (Supplemental Figure 4). Thus, the FLAG-SPL7 transgene is functional in vivo.
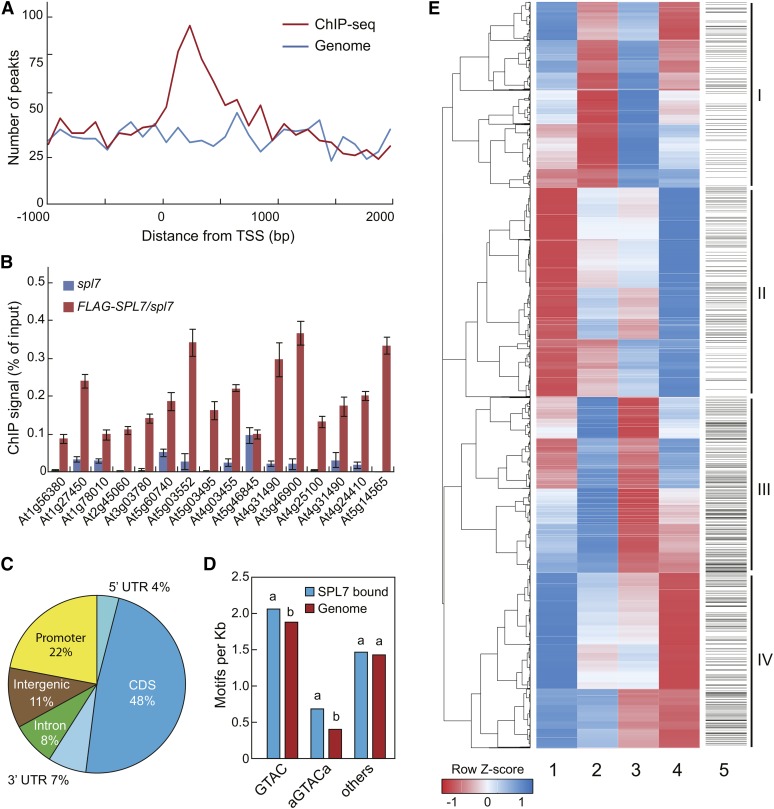
Using 35S:FLAG-SPL7/spl7, we performed anti-FLAG ChIP sequencing and generated 17.8 million reads. As a control, the ChIP sequencing procedure was applied to the spl7 mutant, which yielded 14.8 million reads (Supplemental Table 1). A total of 1535 specific SPL7 binding peaks were identified and found to locate predominantly near the TSS (Figure 2A; Supplemental Data Set 1, sheet 1). For verification, we performed ChIP-qPCR analysis on randomly selected SPL7-occupied regions (Supplemental Figure 5A) and confirmed SPL7 binding for 15 of the 16 tested loci (94%; Figure 2B). The SPL7 binding peaks overlap with 1266 genes, including 10 miRNA genes (Supplemental Figure 5B and Supplemental Data Set 1, sheet 2), whereas 11% of the peaks reside in intergenic regions (Figure 2C). Of the binding sites assigned to genes, most localized to the exons (48%) and the proximal promoter regions (22%; Figure 2C). Gene Ontology (GO) analysis revealed that these genes preferentially associate with GO terms such as “response to stimulus and stress,” “photosynthesis,” “regulation of biological quality,” and “postembryonic development” (Supplemental Figure 5C). Regarding annotated pathways, photosynthesis, carbon fixation, nitrogen metabolism, and glyoxylate and dicarboxylate metabolism are among the most significantly enriched (Supplemental Figure 5D). These results indicate that SPL7 target genes are involved in primary metabolism and responsive to environmental stimuli.
Figure 2.
Genome-Wide Analysis of the SPL7 Regulon.
(A) Distribution of SPL7 binding peaks relative to the TSS. For genes with detected SPL7 binding, the regions 2000 bp downstream and 1000 bp upstream of the TSS were aligned and divided into 30 intervals. The number of genes with SPL7 binding (red) and the number of randomly selected genomic regions (blue) located in each interval were plotted.
(B) ChIP-qPCR validation of 16 randomly selected SPL7 binding sites. ChIP was performed in 35S:FLAG-SPL7/spl7 and spl7 seedlings with the anti-FLAG antibody. SPL7 binding profiles at these loci are depicted in Supplemental Figure 5. Values from quantitative PCR analysis were normalized to their respective DNA inputs. Data are means ± sd (n = 3).
(C) Distribution of SPL7 binding sites across annotated genomic regions. The percentages of binding sites located in the 5′ and 3′ untranslated region (UTR), coding region (CDS), promoter (1-kb region upstream of the TSS), and intergenic region are shown.
(D) Both the GTAC tetranucleotide and the AGTACA/TGTACT hexanucleotide, but none of other related hexanucleotides, are overrepresented in the SPL7 binding sites compared with random genome sequences. The same letters above the columns indicate no statistical difference, while different letters denote groups with significant differences (ANOVA, P < 0.01).
(E) Hierarchical clustering analysis of SPL7-regulated genes. The heat map was generated with differentially expressed genes from four pairwise comparisons: spl7/SC versus wild type/SC (column 1), wild type/DC versus wild type/SC (column 2), spl7/SC versus spl7/DC (column 3), and wild type/DC versus spl7/DC (column 4). Each row represents a gene whose scaled expression value, denoted as the row Z score, is plotted in a color scale with blue indicating higher expression and red indicating lower expression. Grouping of the four major clusters is indicated on the far right. The SPL7 binding pattern is shown as column 5, in which a horizontal line indicates SPL7 binding to a given gene.
SPL7 is known to recognize DNA motifs containing the GTAC sequence (Yamasaki et al., 2009; Sommer et al., 2010). In the identified SPL7 binding sites, this tetranucleotide is significantly overrepresented compared with random genome sequences, based on ANOVA (Figure 2D). Closer inspection revealed that the hexanucleotide in which GTAC is symmetrically flanked by A/T, but not other GTAC-encompassing hexanucleotides, is enriched in SPL7 binding sites (Figure 2D). By EMSA analysis, we confirmed that the A/TGTACT/A motif indeed has stronger affinity for SPL7 than other similar sequences (Supplemental Figure 6A). Moreover, we discovered a novel motif overrepresented in the SPL7-occupied regions that resembles DNA elements recognized by the zinc finger family of transcription factors to which SPL7 belongs (Badis et al., 2008; Supplemental Figure 6B). Binding of SPL7 to this motif was confirmed by EMSA (Supplemental Figure 6C). Furthermore, systematic scanning of the SPL7 binding sites revealed that several other known cis-elements appear at a higher frequency than the genome average (Supplemental Figure 6D). Together, these results indicate that SPL7 recognizes different classes of DNA motifs and acts with other factors to target a broad spectrum of genes.
Next, we conducted whole-transcriptome RNA sequencing using wild-type and spl7 seedlings grown under either DC or SC conditions. We obtained a total of 158 million reads for the four tested samples (Supplemental Table 1) and made four pairwise comparisons, revealing that the expression of 4090 genes is influenced by copper (Supplemental Figure 7A). Clustering analysis revealed that these genes form four major groups (Figure 2E; Supplemental Data Set 1, sheets 3 to 6). Each group has roughly the same numbers of genes (1018 in group I, 1147 in group II, 964 in group III, and 961 in group IV; Supplemental Figure 7B). To validate the RNA sequencing data, we performed RT-qPCR on a handful of copper-responsive as well as randomly selected genes. As shown in Supplemental Figure 8, the results generated by the two methods agreed well.
The four groups of genes exhibit distinctive and contrasting transcriptional profiles. Group I genes have lower levels in the wild type/DC versus wild type/SC comparison and higher levels in the spl7/SC versus spl7/DC comparison, indicating that these genes are induced by the SC condition. Furthermore, most group I genes showed higher levels in the spl7/SC versus wild type/SC comparison but lower levels in the wild type/DC versus spl7/DC comparison (Figure 2E), indicating that SPL7 acts as a negative regulator for these genes. Interestingly, group III genes generally exhibited the opposite behavior to group I genes (Figure 2E), indicating that these genes are induced by DC with SPL7 acting as a positive regulator, although exceptions exist. By contrast, group II genes showed lower levels in the spl7/SC versus wild type/SC comparison, higher levels in the wild type/DC versus spl7/DC comparison, but not much change in the wild type/DC versus wild type/SC and the spl7/SC versus spl7/DC comparisons (Figure 2E), indicating that these genes do not respond to changes in copper regime but require SPL7 to maintain proper levels. Thus, SPL7 acts primarily as a positive regulator of these genes, regardless of the copper condition. Again, the behavior of group IV genes was generally opposite to that of group II (Figure 2E), indicating that the expression of the majority of group IV genes is independent of copper, with SPL7 acting as a negative regulator. Thus, the general trend of Figure 2E revealed that SPL7 could function as either a positive or a negative regulator in the presence or absence of changing copper conditions.
Global analysis showed that the four groups of genes are preferentially associated with different GO terms and annotated pathways, suggesting that SPL7 can broadly modulate primary metabolism and participate in stress responses (Supplemental Figure 7B). Interestingly, group III and IV are about 2-fold enriched with genes bound by SPL7 compared with groups I and II, respectively (Figure 2E; Supplemental Figure 7B). Collectively, analyses of the SPL7 regulon indicate that SPL7 is a global regulator for proper molecular responses to copper regimes and participates in the regulation of other genes through distinct modes of action.
SPL7 and HY5 Coregulate a Large Cohort of Genes
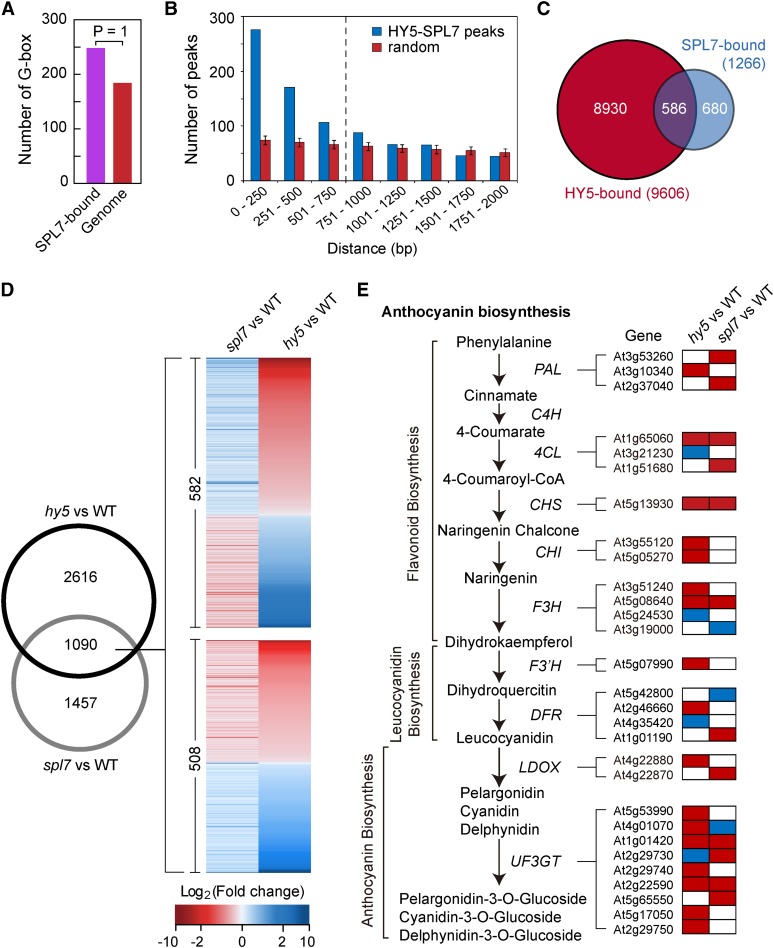
The availability of the SPL7 regulon allowed a comprehensive elucidation of the light-copper crosstalk mediated by the SPL7-HY5 feedback loop. We first sought to identify protein-coding and miRNA genes directly targeted by both SPL7 and HY5 based on the global ChIP data reported here and previously (Zhang et al., 2011; Supplemental Figure 9A). Consistent with their interaction (Figure 1), the G-box, which is recognized by HY5, was found to be overrepresented in the SPL7 binding sites (Figure 3A). This observation prompted us to examine whether SPL7 and HY5 binding sites are clustered. To this end, we performed computational simulations and found that SPL7- and HY5-occupied regions are more likely to locate in close proximity than randomly selected genomic sequences (Figure 3B). Together, these two pieces of evidence support the notion that SPL7 and HY5 bind to a specific set of targets. Using a distance of 750 bp as the threshold, 586 genes, including MIR159a, MIR398b, and MIR408, were identified as common targets of SPL7 and HY5 (Figure 3C; Supplemental Data Set 1, sheet 7). Global analysis revealed that the GO terms “photosynthesis” and “response to stimulus” are most significantly associated with these genes (Supplemental Figures 9B and 9C).
Figure 3.
SPL7 and HY5 Coregulate a Large Cohort of Genes.
(A) The G-box is significantly enriched in the SPL7 binding sites compared with random genomic sequences (posterior probability = 1).
(B) Clustering of SPL7 and HY5 binding sites defined by the global ChIP data. The distance between neighboring SPL7 and HY5 binding sites was calculated, and the number of sites as a function of distance in 250-bp intervals is plotted in blue. The control (red) summarizes 100× simulation of randomly selected genomic loci by the same analysis, with the error bars representing sd. The vertical dashed line indicates a cutoff distance (750 bp) below which SPL7 and HY5 binding sites show significant clustering based on a hypergeometric test (P < 0.001).
(C) Venn diagram showing the overlap of SPL7 and HY5 targeted genes. Using the cutoff illustrated in (B), 586 genes were considered bound by both SPL7 and HY5.
(D) The Venn diagram on the left shows the overlap of SPL7- and HY5-dependent genes under the DC/HL condition. Heat maps on the right illustrate expression changes of the 1090 genes in spl7 and hy5 compared with the wild type. Top, 582 genes influenced by SPL7 and HY5 in the opposite direction; bottom, 508 genes influenced in the same direction.
(E) Anthocyanin biosynthesis as a typical pathway coregulated by SPL7 and HY5. Biochemical steps leading to anthocyanin production are depicted on the left, with genes encoding the relevant enzymes listed. Boxes on the right represent differentially expressed genes in either spl7 or hy5 that are involved in the pathway. Relative expression levels of these genes in either spl7 or hy5 are shaded with different colors, with red indicating reduced expression, blue indicating increased expression, and white indicating no significant change in spl7 or hy5 compared with the wild type.
At the transcript level, SPL7 and HY5 each influence the expression of hundreds of genes, based on RNA sequencing data (Supplemental Data Set 1; Zhang et al., 2011). Comparison of these data revealed that SPL7 and HY5 commonly impact the transcript level of a set of 1090 genes. Interestingly, SPL7 and HY5 modulate 582 of these genes in the opposite direction (upregulated in spl7 but downregulated in hy5 in comparison with the wild type, and vice versa) and 508 genes in the same direction (Figure 3D; Supplemental Data Set 1, sheet 8). Although the exact numbers in this comparison may not be taken literally, as the RNA sequencing experiments were not performed side by side, this result does reveal that SPL7 and HY5 act together to differentially regulate a large cohort of genes. To understand these genes in more detail, we analyzed their associated biochemical pathways.
The production of anthocyanin as part of the flavonoid pathway requires 58 genes that encode the multiple enzymes involved (Solfanelli et al., 2006). According to the RNA sequencing data, 29 of these genes are significantly influenced by either SPL7 or HY5, encompassing essentially every enzymatic step of anthocyanin biosynthesis (Figure 3E). For example, the gene encoding chalcone synthase, the first committed enzyme in flavonoid biosynthesis, showed decreased expression in hy5 and spl7 mutants. Interestingly, because the pathway involves many isozymes that are encoded by members of small paralogous gene families, SPL7 and HY5 appear to work on different family members such that an overall upregulation is achieved. Examples in this regard include genes encoding phenylalanine ammonia lyase, the first committed step in the phenyl propanoid pathway leading to flavonoid biosynthesis, and leucoanthocyanidin dioxygenase, which is involved in proanthocyanin biosynthesis. Thus, a clear trend was observed that SPL7 and HY5 coordinately promote gene expression leading to increased anthocyanin synthesis.
We also analyzed genes related to photosynthesis, which revealed that SPL7 and HY5 both promote the expression of genes involved in the light reactions (Supplemental Figure 10A) but repress genes involved in photorespiration (Supplemental Figure 10B). Interestingly, genes involved in the Calvin cycle are regulated by SPL7 and HY5 in different ways. It appears that genes responsible for the first stage of the cycle, in which a CO2 molecule is incorporated into one of the two three-carbon molecules, are coordinately promoted by SPL7 and HY5. By contrast, genes involved in the regeneration of ribulose-1,5-bisphosphate are not positively regulated by SPL7 and HY5 (Supplemental Figure 10C). Taken together, our results indicate that SPL7 and HY5 are able to exert sophisticated regulation of their target genes involved in various pathways and processes, which presumably constitutes the molecular basis for the light-copper crosstalk.
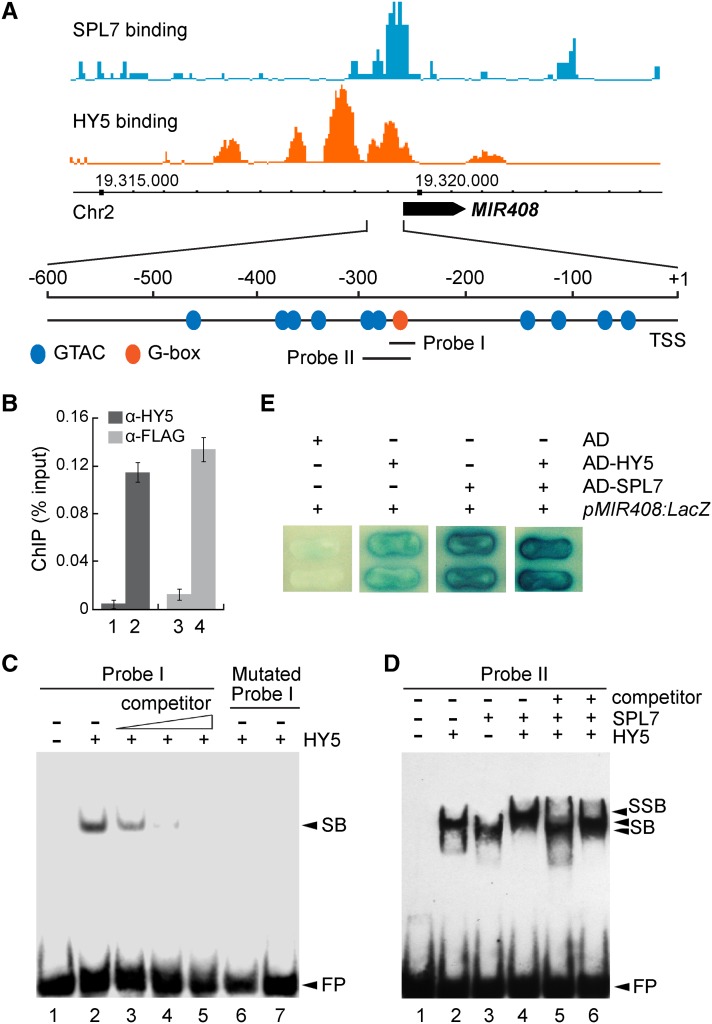
As an example, we tested MIR408, which is encoded by a single locus (At2g47015) in Arabidopsis. Searching the proximal promoter region upstream of the TSS revealed a G-box (CACGTG) and an array of GTAC motifs that clearly coincide with the HY5 and SPL7 binding peaks, respectively (Figure 4A). Previously, we demonstrated through EMSA that SPL7 binds to the GTAC motifs in the MIR408 promoter in vitro (Zhang and Li, 2013). Here, we show through ChIP-qPCR analysis that the anti-FLAG antibody could pull down the MIR408 promoter from 35S:FLAG-SPL7/spl7 but not spl7 plants (Figure 4B). Likewise, the binding of HY5 to the MIR408 promoter was confirmed both in vivo by ChIP-qPCR and in vitro by EMSA (Figures 4B and 4C).
Figure 4.
SPL7 and HY5 Cobind to the MIR408 Promoter.
(A) SPL7 and HY5 occupancy at the MIR408 locus based on global ChIP data, which were mapped onto the Arabidopsis genome coordinates and visualized using the Affymetrix Integrated Genome Brower. The position of pre-miR408 is depicted as a black arrow. Orange and blue ovals in the promoter region represent the GTAC and G-box-like motifs, respectively. Probes used in subsequent EMSA analyses are indicated.
(B) Confirmation of HY5 and SPL7 binding to the MIR408 promoter by ChIP-qPCR analysis. ChIP was performed in the indicated genotypes using either the anti-HY5 or anti-FLAG antibody. Data are means ± sd (n = 3). The numbers 1 to 4 denote hy5, the wild type, spl7, and 35S:FLAG-SPL7, respectively.
(C) EMSA analysis of HY5 binding to the G-box in the MIR408 promoter (probe I in A). Lane 1, labeled probe alone; lane 2, labeled probe incubated with recombinant HY5; lanes 3 to 5, excessive unlabeled probe was added as a competitor with the following competitor:probe ratios: 50 (lane 3), 100 (lane 4), and 200 (lane 5); lanes 6 and 7, a mutated probe (CACGTG changed to CTGCAG) incubated with the same amount of HY5 as in lanes 2 to 5 or eight times more HY5, respectively. FP, free probe.
(D) Cobinding of SPL7 and HY5 to the MIR408 promoter revealed by EMSA. Labeled probe II was incubated without protein (lane 1), with HY5 alone (lane 2), SPL7 alone (lane 3), both HY5 and SPL7 (lane 4), HY5 plus SPL7 and 200-fold excessive mutated probe (CACGTG changed to CTGCAG) as a competitor (lane 5), and HY5 plus SPL7 and 200-fold excess mutated probe (GTAC changed to CATG) as a competitor (lane 6). SB, shift band; SSB, super shift band.
(E) Yeast one-hybrid assay testing the cobinding of SPL7 and HY5 to the MIR408 promoter. Yeast cells containing pMIR408:LacZ were transformed with HY5, SPL7, or HY5 plus SPL7 fused with the Gal4 activation domain (AD) and grown on medium containing X-Gal. Cells expressing the activation domain alone were used as the negative control.
Next, we investigated whether SPL7 and HY5 could bind simultaneously to the MIR408 promoter in vitro. A DNA fragment (−292 to −250) that contains two GTAC motifs and the G-box (Figure 4A) was tested in EMSA for binding with HY5 and SPL7. As shown in Figure 4D, the addition of both SPL7 and HY5 in one reaction produced a supershift band with reduced mobility compared with the band produced by adding either protein alone. When two mutant versions of the probe (one in which the G-box is mutated from CACGTG to CTGCAG and the other in which the two GTAC motifs are both mutated to CATG) were used as competitors, production of the supershifted band was effectively abolished (Figure 4D). Together, these results indicate that SPL7 and HY5 can interact together with the same DNA molecule in vitro. Furthermore, transformation of yeast cells containing the LacZ reporter gene placed under the control of the MIR408 promoter with either HY5 or SPL7 resulted in LacZ expression, whereas additive activation was achieved with the cotransformation of both SPL7 and HY5 (Figure 4E). Together, these data demonstrate that SPL7 and HY5 bind simultaneously to the MIR408 promoter via the GTAC and G-box motifs, respectively.
MIR408 Participates in the Coordinated Copper and Light Response
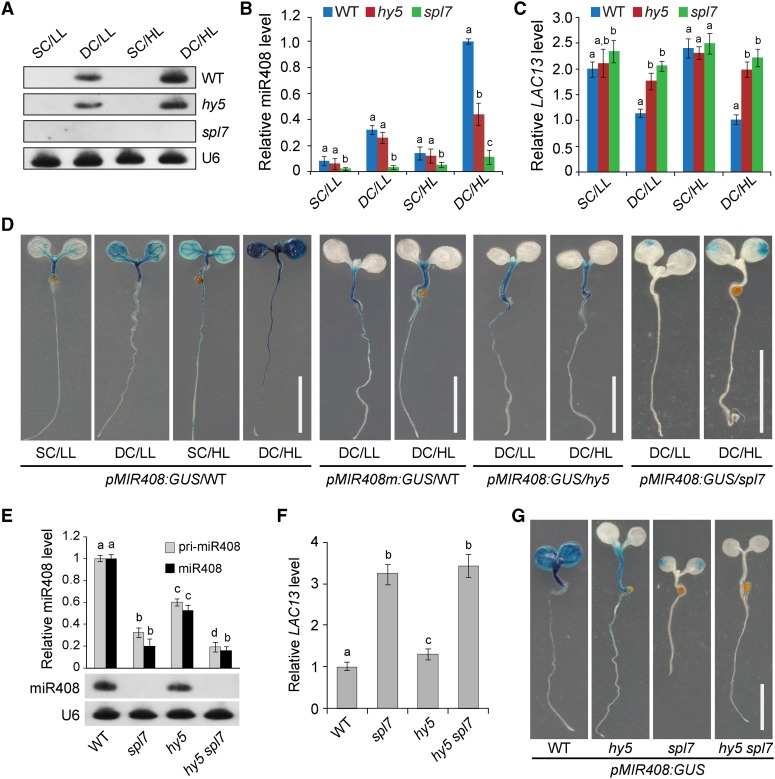
To functionally dissect the SPL7-HY5 network, we first examined miR408 abundance in response to changing levels of copper and light. Both RNA gel blot and RT-qPCR analyses revealed that miR408 is present at the highest level under the DC/HL condition, intermediate under DC/LL and SC/HL, and lowest under SC/LL (Figures 5A and 5B). Additionally, we found that induction of MIR408 by DC/HL requires both SPL7 and HY5, as miR408 accumulation is impaired in the spl7 mutant and partially so in hy5 (Figures 5A and 5B). Previously, the LAC13 gene, which encodes a copper-containing laccase, was identified as a cleavable miR408 target (Abdel-Ghany and Pilon, 2008). Therefore, we investigated whether the expression of LAC13 is affected by growth conditions that impact miR408 abundance. Based on RT-qPCR analysis, we found that copper level and light intensity influence LAC13 in the wild type in a pattern contrary to that of miR408 (Figure 5C). Interestingly, in both spl7 and hy5 mutants, LAC13 became less sensitive to light and copper changes and remained at relatively high levels (Figure 5C). These results indicate that MIR408 and its target gene are regulated by coordinated light and copper signaling.
Figure 5.
HY5 and SPL7 Coordinately Mediate MIR408 Expression in Response to Changing Light and Copper Conditions.
(A) RNA gel blot analysis of miR408 levels in wild-type, hy5, and spl7 seedlings in four different light and copper regimens. U6 small nuclear RNA was used as the loading control.
(B) and (C) RT-qPCR analysis of miR408 levels (B) and the miR408 target gene LAC13 (C) in wild-type, hy5, and spl7 seedlings. For comparison, levels of miR408 and LAC13 in DC/HL were set as 1.
(D) GUS activity in various transgenic plants. The pMIR408:GUS or pMIR408m:GUS reporter was expressed in the wild type or the indicated mutant background. Seedlings grown under different combinations of light and copper conditions were stained for GUS activity and visualized. Bars = 1 cm.
(E) RT-qPCR analysis of miR408 and pri-miR408 (top) and RNA gel blot analysis of miR408 levels (bottom) in wild-type, spl7, hy5, and hy5 spl7 seedlings under the DC/HL condition.
(F) RT-qPCR analysis of LAC13 transcript levels in the indicated genotypes under the DC/HL condition. Levels of miR408 and LAC13 in the wild type were set as 1.
Data for RT-qPCR are means ± sd (n = 3). The same letters indicate no statistical difference, while different letters denote groups with significant differences (ANOVA, P < 0.01).
(G) GUS activity in transgenic plants expressing pMIR408:GUS in the wild-type, spl7, hy5, and hy5 spl7 backgrounds. Bar = 1 cm.
We generated two reporter constructs in which the β-glucuronidase (GUS) gene is fused with the native MIR408 promoter (pMIR408:GUS) or a mutated promoter where the G-box CACGTG was mutated to CTGCAG (pMIR408m:GUS). We then transformed these two reporters into Arabidopsis plants of different genotypes. In wild-type seedlings expressing pMIR408:GUS, GUS activity is strongest under the DC/HL condition, intermediate under DC/LL and SC/HL, and weakest under SC/LL (Figure 5D), consistent with the transcript analyses (Figures 5A and 5B). Furthermore, pMIR408m:GUS generates drastically reduced GUS activity with abolished light responsiveness, which could be phenocopied by expressing pMIR408:GUS in the hy5 background (Figure 5D). When expressing pMIR408:GUS in the spl7 mutant, the GUS activity exhibited a significant decrease, with a weak light responsiveness still remaining in the cotyledons (Figure 5D). These results confirmed that MIR408 activation in response to light and copper is controlled at the transcription level by HY5 and SPL7, respectively.
The GUS staining pattern revealed differences between cotyledons and hypocotyls (Figure 5D). We thus sampled these two organs separately for RT-qPCR analysis and found that the miR408 level is high in wild-type hypocotyls under DC/HL but lower under both DC/LL and DC/HL in cotyledons (Supplemental Figure 11A). In hypocotyls, MIR408 and LAC13 show a much stronger anticorrelation than in cotyledons (Supplemental Figures 11A and 11B). SPL7 is clearly required for MIR408 induction in both cotyledons and hypocotyls under all tested conditions. While this is also the case for HY5 in the cotyledons, it only acts positively for MIR408 expression in the hypocotyls under the HL condition (Supplemental Figure 11A). These observations are generally consistent with GUS activities (Figure 5D), suggesting that environment-induced MIR408 expression is specifically regulated in different organ types, which likely involves additional regulators.
To further analyze the joint effect of HY5 and SPL7 on MIR408, we generated the hy5 spl7 double mutant. RNA gel blot analysis revealed that accumulation of miR408 is essentially abolished in hy5 spl7 seedlings as in spl7 (Figure 5E). RT-qPCR analyses confirmed this result and showed further reduction of pri-miR408 in hy5 spl7 compared with either single mutant (Figure 5E). As expected, LAC13 expression levels generally increased with the decreased levels of miR408 in the mutants (Figure 5F). Finally, we checked the activity of pMIR408:GUS in the hy5 spl7 double mutant and found further diminished GUS staining compared with that in both single mutants (Figure 5G). Together, our results demonstrate that SPL7 and HY5 act additively to control MIR408 transcription in response to varying growth conditions, with SPL7 playing a more dominant role in determining miR408 level.
The SPL7-HY5-MIR408 Circuit Controls Expression Dynamics of Target Genes
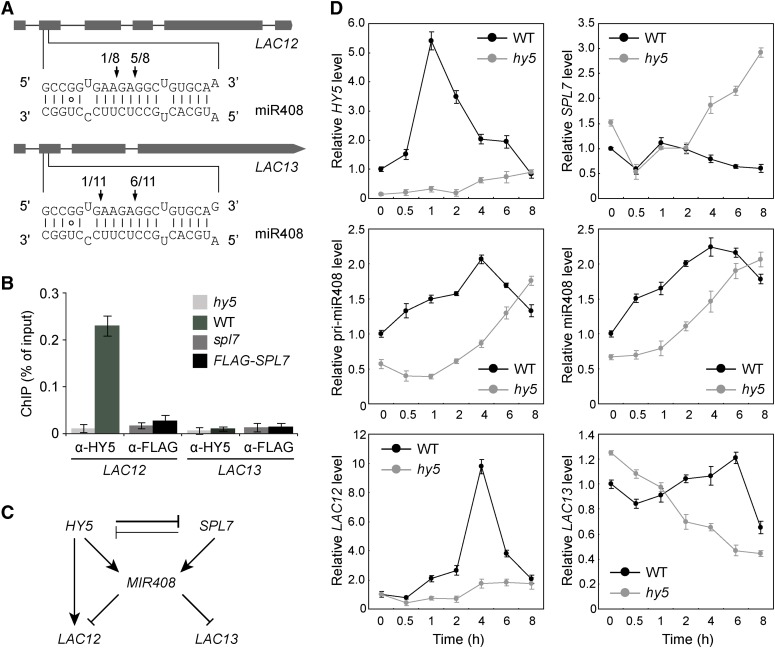
To investigate the molecular role of MIR408 in the SPL7-HY5 network, we began by examining two paralogous miR408 target genes, LAC12 and LAC13, which share essentially identical binding sites for miR408 (Figure 6A). Inspection of the global occupancy data revealed that LAC12 but not LAC13 is targeted by HY5 while neither is directly regulated by SPL7, a pattern also confirmed by ChIP-qPCR analysis (Figure 6B). A diagram summarizing these and the aforementioned results is shown in Figure 6C. According to this diagram, one of the molecular functions of the SPL7-HY5-MIR408 circuit is to differentially regulate LAC12 and LAC13. Therefore, we monitored the expression dynamics of these genes by RT-qPCR analysis in wild-type and hy5 seedlings transitioning from LL to HL.
Figure 6.
The miR408 Target Genes LAC12 and LAC13 Are Differentially Regulated in the HY5-SPL7 Network.
(A) Confirmation of miR408 targeting on LAC12 and LAC13 by 5′ RNA ligase-mediated RACE. Gene structures of LAC12 and LAC13 are shown on the top, with shaded boxes representing exons. The complementary mRNA and miRNA sequences are shown on the bottom. Perfect base pairing is shown as vertical dashes, whereas G:U wobble pairing is indicated by circles. Vertical arrows mark the sequenced cleavage sites with the frequency of clones shown.
(B) Analysis of HY5 and SPL7 binding to the LAC12 and LAC13 promoters by ChIP-qPCR analysis. ChIP was performed in the indicated genotypes using either the anti-HY5 or anti-FLAG antibody. The resultant DNA was analyzed by quantitative PCR with the values normalized to their respective DNA inputs.
(C) Regulatory interactions among HY5, SPL7, MIR408, LAC12, and LAC13. Mutual inhibition between HY5 and SPL7 is based on the molecular data presented in Figure 1. The coordinated transcriptional regulation of MIR408 by HY5 and SPL7 is deduced from Figure 4. The regulation of LAC12 and LAC13 by miR408 at the posttranscriptional level and by HY5 at the transcriptional level is as indicated in (A) and (B), respectively.
(D) Quantitative analysis of HY5, SPL7, miR408, pri-miR408, LAC12, and LAC13 transcript levels in wild-type and hy5 seedlings. Seedlings were grown under the DC/LL condition, transferred to DC/HL at time 0, and assayed by RT-qPCR at the indicated time points thereafter.
After switching from LL to HL, the LAC13 transcript level fluctuates modestly in wild-type seedlings (Figure 6D). However, in hy5, as the miR408 level continues to increase, the LAC13 level declines linearly throughout the time course (Figure 6D), indicating that miR408-mediated posttranscriptional regulation becomes predominant in hy5. By contrast, HY5-based regulation of LAC12 results in a strong transient induction in the wild type and low expression in hy5 (Figure 6D), suggesting that miR408-mediated repression of LAC12 is secondary, although it has identical miR408 binding sites to LAC13. The distinct expression patterns of LAC12 and LAC13 thus indicate that the SPL7-HY5-MIR408 loop is capable of differentially regulating paralogous target genes, providing functional support for its role in executing precise gene expression programs.
It should be noted that the above time-course experiment was done with plants grown under continuous light. Yet, HY5, SPL7, and MIR408 all exhibit pulse-like or rhythmic expression dynamics that peak in a time-dependent manner (Figure 6D), suggesting possible circadian regulation. Meanwhile, analyzing previous global ChIP studies revealed that HY5 targets several key genes in the circadian clock, including CCA1, LHY, LSCB1, and COL1 (Lee et al., 2007; Zhang et al., 2011). In this work, we also found that these genes (except LHY) are targeted by SPL7 (Supplemental Data Set 1, sheet 7). In addition, copper homeostasis has been shown to affect circadian rhythm-related plant growth (Andrés-Colás et al., 2010). Thus, future investigations will be required to elucidate the interplay between circadian rhythms and the HY5-SPL7 circuit.
The SPL7-HY5-MIR408 Loop Controls Plant Development
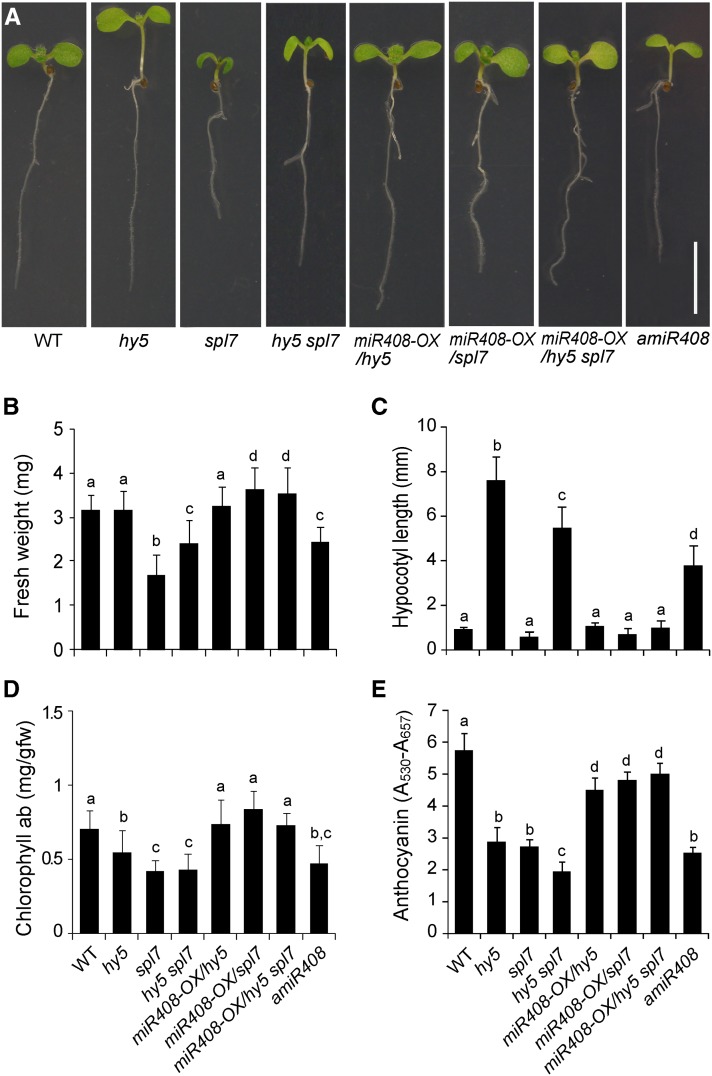
The spl7 and hy5 mutants display distinct phenotypes during early development. When grown in light, the hy5 seedlings have characteristically long hypocotyls while spl7 has reduced growth under DC (Figure 7A; Oyama et al., 1997; Yamasaki et al., 2009). Analyzing the hy5 spl7 double mutant revealed that SPL7 and HY5 interact in different ways to regulate different aspects of development. For example, hy5 spl7 displays intermediate phenotypes in terms of fresh weight under DC (Figure 7B) and hypocotyl length (Figure 7C), phenotypes similar to the single mutants (e.g., chlorophyll content; Figure 7D), and more severe phenotypes such as lower anthocyanin content than the single mutants (Figure 7E). Consistent with a previous report (Yamasaki et al., 2009), we found the spl7 defects could be rescued by SC (Supplemental Figure 12). Interestingly, under this condition, the double mutant still has intermediate hypocotyl length (Supplemental Figure 12C) but no longer displays a more severe phenotype regarding anthocyanin accumulation (Supplemental Figure 12D), but it does have a strong phenotype regarding fresh weight (Supplemental Figure 12E). Thus, SPL7 and HY5 follow distinct genetic modes for controlling diverse aspects of plant development under different growth conditions, which is consistent with the large set of genes regulated by both transcription factors (Figure 3).
Figure 7.
Complementation of the hy5 and spl7 Mutations by MIR408 Overexpression.
(A) Seedling morphology of eight genotypes (as indicated) in which miR408 has varied expression levels. Seedlings were grown under the DC/HL condition and photographed 7 d after germination. Bar = 1 cm.
(B) to (E) Quantitative measurement of fresh weight (B), hypocotyl length (C), chlorophyll (D), and anthocyanin content (E) in seedlings of the eight genotypes. Genotypes labeled with the same letters have no statistical difference, while different letters denote groups with significant differences (ANOVA, P < 0.01). Data are means ± sd from n biological replicates, where n ≥ 30 for (B) and (C) and n = 3 for (D) and (E).
[See online article for color version of this figure.]
To further examine the role of miR408 in development, we employed a previously developed amiR408 line in which a constitutively expressed artificial miRNA (amiR408) silences MIR408 (Zhang and Li, 2013). We found that amiR408 seedlings display elongated hypocotyls and reduced fresh weight and pigment content compared with the wild type (Figure 7). Thus, loss of function in MIR408 impacts all the examined phenotypes. Furthermore, the overall phenotype of amiR408 resembles mostly the hy5 spl7 double mutant (Figure 7), indicating that MIR408 is a critical component of the HY5-SPL7 network. Finally, we sought to test whether overexpression of miR408 could rescue the developmental defects of spl7, hy5, or hy5 spl7. Consistent with previous findings (Zhang and Li, 2013), we found that the 35S:pre-miR408 transgene partially rescues the reduced growth vigor of spl7 under DC (Figure 7). Strikingly, introduction of the 35S:pre-miR408 transgene into the hy5 and hy5 spl7 backgrounds, which results in the accumulation of miR408 and the downregulation of miR408 target genes (Supplemental Figure 13), could also completely or partially rescue all the examined phenotypes (Figure 7).
Sucrose is known to influence the growth and development of Arabidopsis seedlings (Gibson, 2005). To test whether growth defects of the hy5, spl7, and hy5 spl7 mutants, and hence the ability of miR408 to rescue such defects, are dependent on sucrose, we supplemented the growth medium with two concentrations of sucrose. In addition to the experiment reported above (Figure 7) in which 1% exogenous sucrose was used, we also tested 0.1% sucrose (Supplemental Figure 14). Comparing Figure 7 and Supplemental Figure 14 revealed that low sucrose reduced all growth parameters of seedlings except hypocotyl length. However, even with low sucrose supplementation, we found that silencing MIR408 mimics the hy5 spl7 double mutant and that constitutively activated miR408 completely or partially rescues all the examined phenotypes of hy5, spl7, as well as hy5 spl7 (Supplemental Figure 14). These results thus suggest that the action of miR408 in the HY5-SPL7 network is likely independent of sucrose regimens.
In contrast with the drastic phenotypes in seedling development, hy5 plants have moderate defects in later stages (Ang et al., 1998). In our experiments, the hy5 mutant displayed normal fresh weight but reduced pigmentation in juvenile plants (Supplemental Figure 15). Consequently, the hy5 spl7 double mutant displayed phenotypes similar to spl7. Regarding miR408, it is interesting that the amiR408 line still displays phenotypes overall similar to the hy5 spl7 double mutant. Furthermore, overexpression of miR408 could rescue all the examined developmental defects in hy5, spl7, and hy5 spl7 (Supplemental Figure 15). These results indicate that the SPL7-HY5-MIR408 loop is functional beyond the seedling stage.
Cellular Function of miR408
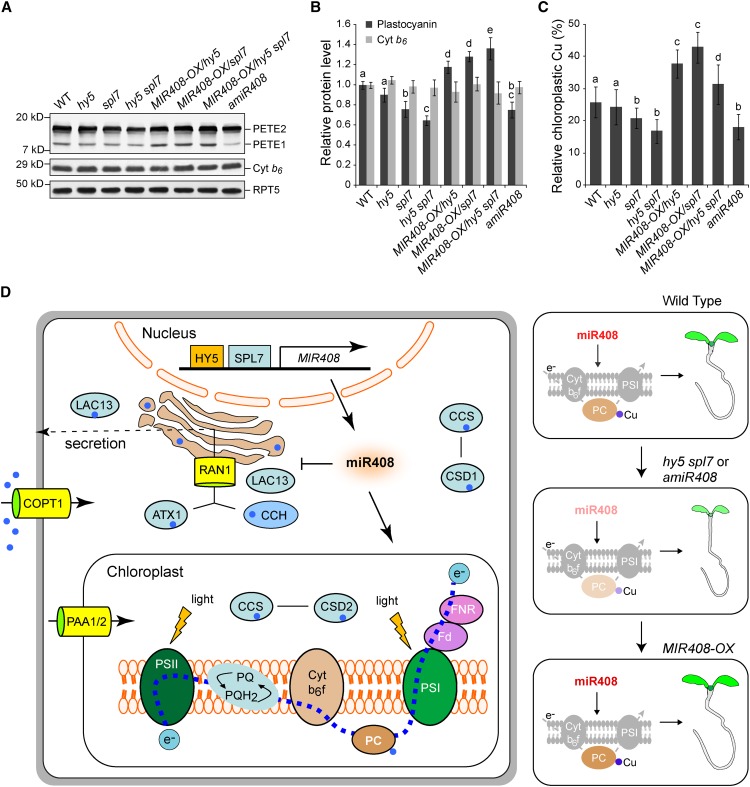
The ability of miR408 to regulate plant growth and chlorophyll level, and to repress copper proteins with nonphotosynthetic usage, prompted us to investigate its involvement in copper allocation and PC abundance. PC is a photosynthetic electron carrier encoded by two paralogous genes, PETE1 and PETE2, in Arabidopsis (Weigel et al., 2003). By means of immunoblotting, we analyzed the levels of PETE1 and PETE2 in various lines in which MIR408 expression is altered. As shown in Figure 8A, the PC antibody detects both PETE1 and PETE2 in seedlings, with PETE2 being the more abundant isoform, as reported previously (Abdel-Ghany, 2009; Pesaresi et al., 2009). Quantification of total PC (PETE1 and PETE2 combined) revealed that its levels are reduced in mutants with impaired MIR408 expression (hy5, spl7, hy5 spl7, and amiR408) and increased when miR408 is constitutively produced (MIR408-OX/hy5, MIR408-OX/spl7, and MIR408-OX/hy5 spl7) compared with the wild type (Figure 8B).
Figure 8.
Cellular Function of miR408.
(A) Detection of PC isoforms in wild-type Arabidopsis seedlings and various genotypes with altered MIR408 expression. Total protein prepared from seedlings grown in the DC/HL condition was fractionated and probed with a commercial PC antibody. Two PC isoforms, PETE1 and PETE2, were detected, with PETE2 being the more abundant isoform. Chloroplast cytochrome b6 (Cyt b6) and RPT5 were used as controls. Blots shown are from one representative of three independent experiments.
(B) Quantification of total PC protein levels in seedlings of various genotypes. The intensity of the bands corresponding to PETE1 and PETE2 was acquired using ImageJ to determine the total PC level, which was then normalized against RTP5 and set to 1 for the wild type. The cytochrome b6 level was also quantified as a control. Data are means ± sd (n = 3).
(C) Relative copper contents in the chloroplast of various genotypes as indicated. Copper content in isolated chloroplasts and whole seedlings of the same genotype was determined separately. Values shown are percentages of chloroplastic copper contents over those of the whole seedlings. Data are means ± sd (n = 4).
Genotypes in (B) and (C) labeled with the same letters have no statistical difference, while different letters denote groups with significant differences (ANOVA, P < 0.01).
(D) A model for SPL7-HY5-regulated MIR408 activation in copper homeostasis and plant development. Depicted on the left are simplified copper transport and utilization pathways that include the main copper transporter COPT1 and the metallochaperones CCS, CCH, and ATX1. These chaperones deliver copper to different internal transporters such as Golgi-localized RNA1 and the chloroplast-specific importers PAA1 and PAA2. Together, the chaperones and transporters mediate copper delivery to specific protein targets such as CSD1 in the cytosol, CSD2 in the chloroplast, PC in the photosynthetic electron transport chain, and LAC13 in the secretory pathway. Elevated miR408, which is promoted by HY5 and SPL7, represses several genes in the copper secretory pathway. Genetically, as shown in the three panels on the right, silencing miR408 expression (amiR408) suppresses PC, reduces chloroplast copper, and compromises seedling development, phenotypes reminiscent of the hy5 spl7 double mutant. Conversely, constitutive action of miR408 in the mutant backgrounds complements such phenotypes. Thus, the cellular function of miR408 is to promote the copper allocation to as well as the abundance of PC, thereby constituting one specific regulatory route downstream of SPL7 and HY5, although its impact on other components of copper homeostasis remains to be determined.
As controls, we examined the protein levels of other photosynthetic electron carriers (Figures 8A and 8B; Supplemental Figure 16). We found that the levels of chloroplast cytochrome b6 and ferredoxin do not change as MIR408 expression alters. Thus, miR408 appears to specifically regulate PC in the photosynthetic electron transport chain. Previous studies have shown that mutations of PETE1 and PETE2 lead to impaired vegetative growth (Weigel et al., 2003; Joliot and Joliot, 2006; Abdel-Ghany, 2009; Pesaresi et al., 2009). In addition, mutations in the transporters that deliver copper to the chloroplast and thylakoid lumen result in drastic reduction of PC and growth defects (Shikanai et al., 2003; Abdel-Ghany et al., 2005). These results suggest that the effects of miR408 levels on plant growth might be exerted through PC abundance.
Because PC is a major copper sink in the chloroplast (Ramshaw et al., 1973), we examined whether miR408-regulated changes in PC abundance are accompanied by corresponding changes in chloroplastic copper levels. To this end, we measured copper content in whole seedlings and found that it does not vary significantly across the examined genotypes (Supplemental Figure 17A). However, compared with the wild type, copper in chloroplasts decreases when MIR408 expression is compromised (hy5, spl7, hy5 spl7, and amiR408) but increases when miR408 accumulates to higher levels (Supplemental Figure 17B). Thus, copper content in chloroplasts as a percentage of total cellular copper tracks the PC level and correlates with miR408 abundance in various genetic backgrounds with varied MIR408 expression (Figure 8C). Together with other data mentioned previously, our results delineate the SPL7-HY5-MIR408 loop as a cellular mechanism for modulating plant growth based on the integration of light and copper signaling (Figure 8D).
DISCUSSION
The regulation of gene expression is fundamental to the integrity and function of all organisms. We now appreciate that complex and sophisticated regulatory networks have evolved in plants for proper gene expression. In addition to transcription factors, miRNAs play crucial roles in the regulatory networks by modulating gene expression at the posttranscriptional level (Voinnet, 2009). In the context of regulatory networks, one of the major challenges to understanding gene expression is to identify and analyze regulatory gene circuits incorporating both transcriptional and posttranscriptional control mechanisms. Such inquires should provide much-needed insights into dynamic gene activity that is critical for plant development and responses to environmental changes.
The SPL7 Regulon in Arabidopsis
Copper is an essential mineral micronutrient and participates as a redox catalytic cofactor in a variety of physiological processes (Pilon et al., 2006; Burkhead et al., 2009). While diminished cellular copper impedes photosynthesis, excessive copper leads to the generation of harmful reactive oxygen species and the replacement of other metal cofactors (Burkhead et al., 2009). Copper homeostasis, therefore, is fundamental to the fitness of plants and involves the expression of genes involved in diverse pathways. As a member of the SBP family of zinc finger transcription factors, SPL7 in Arabidopsis (Yamasaki et al., 2009) and its ortholog CRR1 in C. reinhardtii (Kropat et al., 2005) are regulators for copper homeostasis. Copper inhibits the DNA binding activity of CRR1 and SPL7 and prevents the transcription of specific target genes in vitro (Sommer et al., 2010). These and other observations have led to the proposal that SPL7 is the copper sensor and copper deficiency promotes its binding to the GTAC motif, which in turn transcriptionally activates the target genes (Yamasaki et al., 2009; Beauclair et al., 2010; Sommer et al., 2010; Bernal et al., 2012).
In this study, we fully elucidated the SPL7 regulon in Arabidopsis. By whole-genome ChIP sequencing, we identified 1535 high-confidence SPL7-bound genomic regions aligned with 1266 gene loci (Figure 2; Supplemental Figure 5 and Supplemental Data Set 1, sheets 1 and 2). A strong enrichment of SPL7 binding in the proximity of the TSS of target genes was observed (Figure 2A), suggesting that distribution of the recognition sites for SPL7 in the genome is highly selective. As a known recognition motif for SPL7 (Birkenbihl et al., 2005; Kropat et al., 2005; Yamasaki et al., 2009; Sommer et al., 2010), the GTAC tetranucleotide, particularly in the context of A/TGTACT/A, is overrepresented in the SPL7-occupied regions (Figure 2D; Supplemental Figure 6A). Additionally, a novel motif in the SPL7 binding sites that resembles DNA elements recognized by zinc finger transcription factors was found (Supplemental Figures 6B and 6C; Badis et al., 2008). Moreover, the SPL7 binding sites are also enriched with recognition motifs for other transcription factors such as the G-box (Figure 3A; Supplemental Figure 6D). These results clearly demonstrate that SPL7 is well connected in the regulatory network through recognizing different classes of DNA motifs.
By means of RNA sequencing, the transcription of ∼4000 genes appears to be influenced by SPL7 under both DC and SC conditions (Figure 2E; Supplemental Figure 7 and Supplemental Data Set 1, sheets 3 to 6), including many involved in primary metabolism, as others have noted (Bernal et al., 2012). Clustering analysis showed that the SPL7-dependent genes form four groups with distinct transcriptional behavior (Figure 2E; Supplemental Figure 7B). Previously, Yamasaki et al. (2009) reported that SPL7 positively regulates several MIR genes, while Bernal et al. (2012) noted that SPL7 represses many metabolism-related genes. Our results indicate that SPL7 could function either as a positive or negative regulator, which is consistent with the analysis of CRR1 in C. reinhardtii (Moseley et al., 2002). Combining the ChIP and RNA sequencing data revealed that half of the SPL7-bound genes (634 of 1266) exhibit SPL7-dependent expression (Supplemental Figure 7B). Although this proportion is much higher than the genome average, SPL7 binding to many target genes may not be sufficient to cause changes in their expression levels. Conversely, a majority of the genes for which proper expression is dependent on SPL7 are not directly bound by the transcription factor (Supplemental Figure 7B). Together, these results suggest that SPL7 controls its regulon through interwoven subprograms coordinated with other transcriptional regulators.
The HY5-SPL7 Interaction Defines a Light-Copper Crosstalk
Combinatorial control is a major mechanism underlying transcriptional regulation in eukaryotes (Carey, 1998). An increasing number of studies in plants have documented combinatorial control involving transcription factors that are implicated in multiple biological processes, such as pigment biosynthesis, carbon metabolism, hormonal responses, organ development, and plant immunity (Mol et al., 1996; Schultz et al., 1998; Hobo et al., 1999; Yanagisawa, 2000; Lara et al., 2003; Shin et al., 2007; Liu and Howell, 2010; Moore and Goldberg, 2011; Park et al., 2011). Enrichment of various DNA elements in SPL7-occupied regions (Figure 3A; Supplemental Figure 6D) indicates that much of the regulatory function of SPL7 is likely fulfilled together with other transcription factors.
Given that copper is essential for photosynthesis (Pilon et al., 2006; Burkhead et al., 2009), it is not surprising, but nevertheless interesting, to find that SPL7 interacts with HY5 in Arabidopsis (Figures 1A to 1C). Because they each impact thousands of genes, the SPL7-HY5 interaction thus defines a previously unknown transcriptional level light-copper crosstalk. Indeed, through global comparison of the SPL7 and HY5 regulons, we show that the SPL7-HY5 feedback loop regulates a large cohort of genes (Figure 3; Supplemental Figures 9 and 10), indicating that the light-copper crosstalk is extensive. Our results further demonstrate that the interplay of the SPL7-HY5 network is performed through different molecular mechanisms. On the one hand, HY5 binds directly to the G-box-like motifs in the SPL7 promoter and represses its expression (Figures 1D and 1E; Supplemental Figure 3). Thus, one aspect of the crosstalk is indirect attenuation of the SPL7 regulon achieved through HY5-mediated SPL7 repression, presumably to put a counterweight on copper-responsive genes based on input from light signaling.
On the other hand, interaction between the two transcription factors entails that they exert combinatorial control over their commonly regulated genes. Because HY5 and SPL7 both can serve as positive as well as negative regulators, the net impact of these two factors on the target genes could be cooperative or antagonistic. In support of this notion, through RNA sequencing, we found that SPL7 and HY5 commonly regulate 1090 genes, with roughly half of these genes being regulated in the same direction by the two transcription factors and half in the opposite direction (Figure 3D; Supplemental Data Set 1, sheet 8). Furthermore, genes coregulated by SPL7 and HY5 are involved in processes such as photosynthesis (Supplemental Figure 10) and the biosynthesis of anthocyanins (Figure 3E), which help to absorb blue-green light and thereby protect photosynthetic tissues from photoinhibition (Feild et al., 2001; Neill and Gould, 2003; Hughes et al., 2005; Solfanelli et al., 2006; Merzlyak et al., 2008). These findings are consistent with observations that chlorophylls and anthocyanins accumulate to high levels under the SC/HL condition (Supplemental Figure 2). Therefore, photosynthesis appears to be a major point of convergence of light signaling through HY5, which perceives solar energy available for harvesting, and copper sensing through SPL7, which is tied to copper allocation to the electron transport chain and the cycling of reactive oxygen species (Burkhead et al., 2009). The sophisticated combinatorial regulations exerted by the HY5-SPL7 loop thus likely constitute one of the molecular mechanisms for mediating the light-copper crosstalk.
As a specific example for functional study, we show that coordinated SPL7-HY5 regulation represents a mechanism for calculated miR408 accumulation, with SPL7 playing a more dominant role in determining miR408 levels (Figures 4 and 5). This mechanism ensures that the miR408 level is low when sufficient copper is present, intermediate in CD and LL conditions, and high when copper is low but light is strong (Figure 5). Thus, transcriptional regulation of MIR408 by both HY5 and SPL7 allows distinct temporal and spatial expression dynamics to be established by combining different input signals. Significantly, we demonstrate that MIR408 is sufficient to activate the HY5-SPL7 network, as constitutively expressed miR408 could rescue or partially rescue all the examined developmental defects of the hy5, spl7, and hy5 spl7 mutants (Figures 7 and 8; Supplemental Figures 14 and 15).
Complementation of the upstream regulators by MIR408 is an intriguing finding because all validated miR408 target genes encode copper proteins designated to the extracellular space (Yamasaki et al., 2007; Abdel-Ghany and Pilon, 2008), which suggests a primary role for MIR408 in copper homeostasis. Our finding indicates that MIR408 is an integral and critical component of the HY5-SPL7 network and provides a specific signaling route that connects the transcriptional and posttranscriptional gene regulatory branches. One implication of this finding is that there are multiple parallel routes for regulatory information flow in the HY5-SPL7 network and that the activation of individual routes is sufficient to excite the network. In addition to MIR408, SPL7 and HY5 coregulate two more miRNAs and 28 transcription factors. It is plausible that the HY5-SPL7 feedback outputs calculated gene expression programs through clustered regulatory loops involving other gene regulators. Further elucidating the interplay among genes in the HY5-SPL7 network should provide much-needed insights into the gene batteries that underpin plant development in response to changing environments.
A Model for HY5-SPL7-Regulated MIR408 Activation in Plant Growth
Based on several lines of evidence, MIR408 is clearly a critical component of the HY5-SPL7 network. Physiologically, relative copper content in the chloroplast decreased when MIR408 expression was undermined, as in hy5, spl7, hy5 spl7, and amiR408 plants, while constitutive activation of MIR408 was sufficient to reverse such decreases (Figure 8C). Molecularly, the abundance of PC, which is a key component of the photosynthetic electron transport chain and the major copper sink in chloroplast (Marschner, 2002; Burkhead et al., 2009), is reduced as a consequence of compromised MIR408 activation but restored by miR408 overproduction (Figures 8A and 8B). Consequently, the cellular contents of chlorophyll (Figure 7D; Supplemental Figures 14D and 15C), which is an approximation of the amount of photons harvested (Maxwell and Johnson, 2000), and anthocyanin (Figure 7E; Supplemental Figures 14E and 15D), which is modulated by the redox status of the plastoquinone pool of the electron transport chain (Das et al., 2011), are reduced in mutants and amiR408 plants but elevated by constitutively expressing miR408.
Reconciling all observations, we propose that transcriptional regulation of MIR408 by HY5 and SPL7 constitutes a mechanism for integrating light and copper signals to control photosynthesis (Figure 8D). In this model, the elevated miR408 level coordinately promoted by HY5 and SPL7 in the DC/HL condition would reduce copper usage in the extracellular space by repressing transcripts encoding copper proteins such as laccases and plantacyanin (Figure 6A; Supplemental Figure 13B). This would result in a preferential allocation of cellular copper to the chloroplast and thus PC (Figures 7D, 7E, and 8A to 8C; Supplemental Figures 14 to 17). We hypothesize that increased copper delivery to PC drives up its expression, which in turn would increase the flux of photosynthetic electron transport, as evident by the chlorophyll content. Conversely, compromised MIR408 activation would deprive a plant of the ability to preferentially deliver copper to PC in all light and copper regimes. This would result in diminished photosynthetic electron flux due to reduced PC and pigmentation.
Given the critical function of PC in vegetative growth (Weigel et al., 2003; Joliot and Joliot, 2006; Abdel-Ghany, 2009; Pesaresi et al., 2009), an inference is that miR408 levels should correlate with the vigor of plant growth. This is indeed the case, as we have shown previously that constitutive activation of MIR408 results in enhanced vegetative growth while silencing MIR408 causes impaired growth (Zhang and Li, 2013). Furthermore, we found that the hy5, spl7, and hy5 spl7 mutants display defects in vegetative development that could be rescued by constitutively activated MIR408 (Figure 7). Given the inherent complexity of copper homeostasis (Figure 8D), it is not yet clear how miR408-based regulation works coordinately with the web of copper chaperones and transporters for economic distribution of this critical transition metal under varying growth conditions. For example, MIR408-OX apparently could increase total copper content in spl7 plants (Supplemental Figure 17A). This implies that miR408 may enhance the expression or activity of copper transporters such as COPT1 and COPT2 in the absence of a functional SPL7, although the signaling mechanism is elusive. Nevertheless, our results collectively indicate that the cellular function of miR408 is to promote copper allocation to the chloroplast and the abundance of PC, thereby constituting one specific regulatory route downstream of SPL7 and HY5 to modulate vegetative growth based on light and copper inputs (Figure 8D).
It should be noted that MIR408 is among the most conserved miRNA families in land plants (Axtell and Bowman, 2008), suggesting that its role in mediating light-copper crosstalk is fundamental to plants. Thus, the SPL7-HY5-MIR408 circuit represents a potentially conserved determinant for photosynthetic activity and hence plant growth and adaptation in a changing environment. Photosynthesis in plants is a relatively inefficient process, with far below 10% of the received solar energy being converted to chemical energy (Zhu et al., 2010). This relatively low efficiency has provided an impetus for researchers to genetically modify plants to achieve greater efficiencies and enhanced growth. So far, much of this effort has centered on carbon fixation (Kirschbaum, 2011). Our finding suggests that manipulating the capacity of the electron transport chain mediated by the SPL7-HY5-MIR408 circuit is an alternative strategy that warrants further investigation.
METHODS
Plant Materials and Growth Conditions
Wild-type plants used were the Arabidopsis thaliana ecotype Columbia-0. Mutants defective in HY5 and SPL7 were hy5-215 (Oyama et al., 1997) and the T-DNA insertion line SALK_093849 (Yamasaki et al., 2009), respectively. To obtain the 35S:FLAG-SPL7/spl7 line, N-terminal FLAG-tagged SPL7 was generated by PCR using a forward primer containing the FLAG sequence, inserted into the pJim19 binary vector under the control of the 35S promoter, and transformed into the spl7 background. Transgenic plants were selected with 20 mg/L Basta. T3 generation homozygous lines were used for all experiments. Transgenic plants expressing the GUS reporter were generated as described previously (Zhang and Li, 2013). Briefly, a mutated miR408 promoter was obtained from the native promoter sequence by bridge PCR to change the CACGTG sequence of the G-box to CTGCAG, cloned into the pCAMBIA-1381Xa vector (CAMBIA), and transformed into various genetic backgrounds as indicated. For each transgene, at least three independent lines were selected with 25 mg/L hygromycin. T2 generation plants of representative lines were used for histochemical staining of GUS activity as described previously (Zhang and Li, 2013). The hy5 spl7 double mutant was generated by crossing hy5-215 and spl7. F2 progeny homozygous for both alleles were identified by PCR analysis of genomic DNA for the presence of the T-DNA and the hy5 allele. This procedure was repeated for the F3 generation, in which a single hy5 spl7 double mutant line was selected and used for all subsequent analyses. The 35S:pre-miR408 transgene, which contains the sequence encompassing the pre-miR408 stem-loop structure (Zhang and Li, 2013), was used to overexpress miR408 in various genetic backgrounds. Transgenic plants were selected with Basta, and T2 generation homozygous lines were used for subsequent experiments.
To grow Arabidopsis seedlings, seeds were surface-sterilized and plated on agar-solidified Murashige and Skoog medium including 0.1% or 1% sucrose and the indicated concentrations of CuSO4. The plates were incubated at 4°C for 4 d in the dark, then transferred to continuous white light with intensity of either 40 or 170 µmol m−2 s−1, and allowed to grow at 22°C for 7 d or other indicated lengths of time. To obtain adult plants, seedlings were transferred to soil and maintained in a growth chamber with the following settings: standard long-day (16 h of light/8 h of darkness) conditions, light intensity of ∼120 µmol m−2 s−1, 50% RH, and temperature at 22°C. All phenotypic characterization experiments were conducted on multiple biological samples and repeated at least three times. Results from one representative experiment are shown in the figures.
ChIP Sequencing
Chromatin isolation was performed with whole seedlings of 35S:FLAG-SPL7/spl7 and spl7 grown under the DC/HL condition according to the procedure described by Bowler et al. (2004). The resuspended chromatin pellet was sonicated at 4°C with a Diagenode Bioruptor set at high intensity for 10 min (30-s on/30-s off intervals). Chromatin was immunoprecipitated with monoclonal anti-FLAG antibody (Sigma-Aldrich) according to the Affymetrix Chromatin Immunoprecipitation Assay Protocol Rev.3. The precipitated DNA (one biological replicate for each sample) was sequenced using the HiSeq 2000 system (Illumina) according to the manufacturer’s instructions. Sequencing reads of 100 bp were mapped to the TAIR10 genome release of Arabidopsis using Bowtie (Langmead et al., 2009), allowing two mismatches and no gaps. Only uniquely mapped reads were retained for further analysis. MACS (Zhang et al., 2008) with customized parameters (bandwidth = 300 bp; P = 1e−05; mfold = 10 to 50; no lambda) was used to call peaks representing enriched SPL7 binding specifically in 35S:FLAG-SPL7/spl7 but not spl7.
We used Multiple Em for Motif Elicitation (Bailey et al., 2006) to identify sequence motifs overrepresented in SPL7 binding sites. To search for the presence of other transcription factor binding sites, a position weight matrix method based on experimentally validated data derived from the Arabidopsis Promoter Binding Element Database (http://exon.cshl.org/cgi-bin/atprobe/atprobe.pl) and the Arabidopsis Gene Regulatory Information Server data set (Davuluri et al., 2003) was used as described previously (Zhao et al., 2013). Posterior probability was calculated using 10,000 times Monte Carlo simulation in MATLAB.
RNA Sequencing
Total RNA from wild-type and spl7 seedlings grown under DC and SC conditions was isolated using the RNeasy Plus Mini Kit (Qiagen). Library construction and sequencing on the HiSeq 2000 platform were performed according to the manufacturer’s instructions (Illumina). One biological replicate for each sample was analyzed. The resultant 100-bp reads were aligned to the TAIR10 genome using TopHat (Trapnell et al., 2009), allowing two mismatches and maximal intron size of 2 kb. Only uniquely mapped reads were used for subsequent analysis. Differentially expressed genes were identified using Cufflinks (Trapnell et al., 2010) with the following parameters: minimal number of alignment = 50, quartile normalization, as false discovery rate < 0.01, and P < 0.05. Results from four pairwise comparisons (spl7/SC versus wild type/SC, wild type/DC versus wild type/SC, spl7/SC versus spl7/DC, and wild type/DC versus spl7/DC) were included for hierarchical clustering analysis based on Pearson’s correlation. For a given gene, the value for each comparison was set to the logarithm of fold change of normalized read counts (per kb transcript per million mapped reads). A heat map of the differentially expressed genes (rows) across the four comparisons (columns) was generated using the row Z score. For each row in a column, the Z score was calculated by subtracting that gene’s mean relative expression level across the four comparisons from its value in that particular comparison and then dividing by the sd across all the comparisons. GO analysis was performed using BiNGO (Maere et al., 2005). ChIP and RNA sequencing data are available in the Gene Expression Omnibus database under accession number GSE45213.
Yeast Assays
For yeast two-hybrid assays, the full-length SPL7 open reading frame was amplified by RT-PCR from wild-type plants and cloned into the B42AD vector (Clontech) to generate the B42AD-SPL7 construct. The LexA-HY5 and B42AD-COP1 constructs were obtained as described previously (Ang et al., 1998). The respective combinations of B42AD and LexA fusion constructs were cotransformed into the yeast strain EGY48 containing the reporter plasmid p8op:LacZ (Clontech). For yeast one-hybrid assay, plasmids for activation domain fusions (AD-SPL7 and AD-HY5) were cotransformed with the LacZ reporter driven by the MIR408 promoter as described (Zhang and Li, 2013) into the yeast strain EGY48. Transformants were grown on proper dropout plates containing X-Gal for blue color development. Yeast transformation and liquid assay were conducted as described in the Yeast Protocols Handbook (Clontech).
Protein Analyses
Protein extraction and immunoblotting were performed as described previously (Feng et al., 2004). The blots were probed with different primary antibodies as follows: anti-FLAG (GenScript), anit-HY5 (Osterlund et al., 2000), anti-RPT5 (Kwok et al., 1999), anti-His (Qiagen), anti-plastocyanin, anti-cytochrome b6, anti-cytochrome oxidase subunit II, and anti-ferredoxin (Acris Antibodies). All immunoblotting experiments were repeated at least three times, with blots from one representative experiment shown in the figures. For the coimmunoprecipitation experiment on the in vivo binding between SPL7 and HY5, total protein extracts were prepared from 35S:FLAG-SPL7/spl7 transgenic seedlings. The anti-FLAG or anti-HY5 antibody was added to the protein extracts and precipitated with Protein A agarose beads (Sigma-Aldrich) following a method described previously (Feng et al., 2004). The precipitates and total extracts were then subjected to immunoblot analysis with antibodies against HY5, FLAG, or RPT5. For in vitro binding, 2 μg of purified recombinant bait proteins (GST-HY5 and GST) and 2 μg of prey protein (6xHis-SPL7) were added to 1 mL of binding buffer containing 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 0.6% Triton X-100. After incubation at 4°C for 2 h, Glutathione Sepharose 4B beads (Amersham Biosciences) were then added and incubated for another 1 h. After washing three times with the binding buffer, pulled down proteins were eluted in 2× SDS loading buffer at 95°C for 10 min, separated on SDS-PAGE gels, and detected by immunoblotting using the anti-His antibody.
RNA Analyses
Total RNA was extracted using the TRIzol reagent (Invitrogen) as suggested by the manufacturer to include the low-molecular-weight fraction of RNA. For RT-qPCR quantification of protein-coding genes and pri-miR408, DNaseI-treated RNA was reverse transcribed using SuperScript II reverse transcriptase (Invitrogen). The resultant cDNA was analyzed using the SYBR Green master mix with the ABI 7500 Fast Real-Time PCR System (Applied Biosystems) in triplicate. The ACTIN7 amplicon was used for normalization. For RT-qPCR quantification of mature miR408, poly(A) tailing and first-strand cDNA synthesis were performed using the NCode miRNA First-Strand cDNA Kit (Invitrogen). A miR408-specific forward primer (complementary to mature miR408) and a universal reverse primer supplied by the manufacturer were used. 5S ribosome RNA was used for normalization. Determination of relative gene expression level was performed using the standard 2−ΔΔC(T) method. All PCR experiments were performed on three independent biological samples, with each including three technical replicas. The experiments were repeated at least three times, and data from one representative experiment are shown in the figures. RNA gel blot analyses of miRNA were performed as described previously (Zhang and Li, 2013) and repeated at least three times, with blots from one representative experiment shown in the figures. Primer and probe sequences are listed in Supplemental Table 2.
EMSA
Full-length HY5 was amplified by RT-PCR and cloned into the vector pET-28a(+) (Novagen). The resulting plasmid was introduced into Escherichia coli BL-21, and His-tagged HY5 was purified with the Ni-NTA Agarose system (Qiagen). Recombinant SPL7 was prepared as described previously (Zhang and Li, 2013). EMSA was performed using digoxigenin-labeled probes and the second-generation DIG Gel Shift Kit (Roche) according to the manufacturer’s instructions. Sequences of probes and primers used in this study are shown in Supplemental Table 2.
Chlorophyll, Anthocyanin, and Glucose Measurements
Measurement of chlorophyll and anthocyanin was performed as described previously (Chory et al., 1989). Briefly, 7-d-old seedlings were harvested, weighed, and homogenized in liquid nitrogen. Chlorophyll a/b was extracted into 80% acetone and quantified as micrograms per gram fresh weight using MacKinney’s specific absorption coefficients, for which chlorophyll a = 12.7(A663) − 2.69(A645) and chlorophyll b = 22.9(A645) − 4.48(A663). For anthocyanin, homogenized samples were incubated overnight in 0.3 mL of 1% HCl in methanol at 4°C and extracted using an equal volume of chloroform after the addition of 0.2 mL of water. The quantity of anthocyanins was determined by spectrophotometric measurement of the aqueous phase (A530 − 0.25×A657) and normalized to the fresh weight of each sample. Measurement of glucose content was performed using the Glucose and Sucrose Assay Kit (Biovision) according to the manufacturer’s instructions. Seedlings were homogenized in a glucose assay buffer, and the supernatant was collected after centrifuging at 12,000 rpm for 10 min. The reaction system was set up in a 100-μL total volume including 50 μL of sample in glucose assay buffer and 50 μL of glucose assay mix (46 μL of glucose assay buffer, 2 μL of glucose probe, and 2 μL of glucose enzyme mix) and incubated at 37°C for 30 min. The A570 was collected, and glucose concentrations of the test samples were calculated based on the standard curve. These experiments were performed on three independent biological samples, as indicated in the corresponding figure legends. The experiments were repeated at least three times, and data from one representative experiment are shown in the figures.
Measurement of Cellular Copper
Harvested 10-d-old seedlings of various genotypes were weighed, then washed twice with 1 mM EDTA and once with double-distilled water. The materials were then desiccated, digested in 1% nitric acid, and used directly for copper analysis with inductively coupled plasma-atomic emission spectroscopy as described previously (Cohu and Pilon, 2007). Chloroplasts were isolated from shoots of seedlings as described previously (Kubis et al., 2008). Briefly, plant tissues were homogenized in the chloroplast isolation buffer (0.3 M sorbitol, 5 mM MgCl2, 5 mM EGTA, 5 mM EDTA, 20 mM HEPES/KOH, pH 8.0, and 10 mM NaHCO3). The homogenate was filtered through two layers of Miracloth and centrifuged at 3000 rpm in the Sorvall RC6 centrifuge with an SLA-1500 rotor for 5 min to pellet the crude chloroplast. To separate the intact chloroplasts from broken chloroplasts and other debris, the pellet was further purified on a continuous 50% (v/v) Percoll gradient through centrifugation at 7000 rpm in the Sorvall RC6 centrifuge with an HB-6 rotor for 10 min. The lower green band in the gradient, which contains intact chloroplasts, was collected. The intactness of the chloroplasts was assessed by phase-contrast light microscopy, and only samples with more than 80% intact chloroplasts were retained. Isolated chloroplasts were then dried, digested in 1% nitric acid, and assayed for copper content. Copper measurements on all genotypes were conducted on four independently prepared biological samples.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MIR408 (At2g47015), SPL7 (At5g18830), HY5 (At5g11260), LAC12 (At5g05390), LAC13 (At5g07130), PETE1 (At1g76100), PETE2 (At1g20340), FD1 (At1g10960), PETB (ATCG00720), and ACTIN7 (At5g09810). The T-DNA insertion mutant used is spl7 (SALK_093849).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Effect of Copper Regimen on the Expression of Copper-Responsive Genes in Arabidopsis.
Supplemental Figure 2. Analysis of the Light-Copper Crosstalk in Arabidopsis.
Supplemental Figure 3. HY5 Directly Binds to the SPL7 Promoter.
Supplemental Figure 4. Complementation of spl7 by the 35S:FLAG-SPL7 Transgene.
Supplemental Figure 5. Analysis of SPL7-Targeted Genes.
Supplemental Figure 6. Identification and Validation of Enriched DNA Elements in the SPL7 Occupied Sites.
Supplemental Figure 7. Global Analysis of SPL7-Regulated Genes.
Supplemental Figure 8. Validation of RNA Sequencing Data by RT-qPCR.
Supplemental Figure 9. Analysis of Genes Cobound by SPL7 and HY5.
Supplemental Figure 10. SPL7 and HY5 Coordinately Regulate Photosynthesis.
Supplemental Figure 11. Expression of MIR408 and LAC13 in Cotyledons and Hypocotyls.
Supplemental Figure 12. Effects of High Copper on the hy5 spl7 Double Mutant.
Supplemental Figure 13. Functional Analysis of miR408.
Supplemental Figure 14. Phenotypes of Seedlings Grown with Low Sucrose.
Supplemental Figure 15. Growth Phenotypes of Adult Plants with Altered MIR408 Levels.
Supplemental Figure 16. Expression of Photosynthetic Electron Carrier Proteins in Seedlings with Varied miR408 Levels.
Supplemental Figure 17. Copper Content in Whole Seedlings and the Chloroplast Fractions of Various Genotypes with Altered MIR408 Expression.
Supplemental Table 1. Summary of ChIP Sequencing and RNA Sequencing Data.
Supplemental Table 2. Oligonucleotide Sequences for the Primers and Probes Used in This Study.
Supplemental Data Set 1. List of Genes for ChIP Sequencing and RNA Sequencing Data.
Supplementary Material
Acknowledgments
We thank Lin Pu at the University of Virginia and Hang He at Peking University for assistance with copper measurement and statistical analysis, respectively. This work was supported by the Peking-Tsinghua Center for Life Sciences (to L.L.), the National Science Foundation (Grant DBI-0922526 to L.L.), and the Helen Hay Whitney Foundation (to H.C.).
AUTHOR CONTRIBUTIONS
H.Z. and L.L. designed the research. H.Z., J.L., and H.C. performed the research. X.Z., H.Z., and L.L. analyzed the data. H.Z., X.W.D., and L.L. wrote the article.
Glossary
- ChIP
chromatin immunoprecipitation
- PC
plastocyanin
- miRNA
microRNA
- HL
high light
- LL
low light
- SC
sufficient copper
- DC
deficient copper
- TSS
transcription start site
- EMSA
electrophoretic mobility shift assay
- ChIP-qPCR
quantitative PCR analysis following chromatin immunoprecipitation
- RT-qPCR
reverse transcription coupled with quantitative PCR
- GO
Gene Ontology
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Abdel-Ghany S.E. (2009). Contribution of plastocyanin isoforms to photosynthesis and copper homeostasis in Arabidopsis thaliana grown at different copper regimes. Planta 229: 767–779. [DOI] [PubMed] [Google Scholar]
- Abdel-Ghany S.E., Pilon M. (2008). MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 283: 15932–15945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Ghany S.E., Müller-Moulé P., Niyogi K.K., Pilon M., Shikanai T. (2005). Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 17: 1233–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés-Colás N., Perea-García A., Puig S., Peñarrubia L. (2010). Deregulated copper transport affects Arabidopsis development especially in the absence of environmental cycles. Plant Physiol. 153: 170–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang L.H., Chattopadhyay S., Wei N., Oyama T., Okada K., Batschauer A., Deng X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1: 213–222. [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Bowman J.L. (2008). Evolution of plant microRNAs and their targets. Trends Plant Sci. 13: 343–349. [DOI] [PubMed] [Google Scholar]
- Badis G., et al. (2008). A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell 32: 878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Williams N., Misleh C., Li W.W. (2006). MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34: W369–W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauclair L., Yu A., Bouché N. (2010). MicroRNA-directed cleavage and translational repression of the copper chaperone for superoxide dismutase mRNA in Arabidopsis. Plant J. 62: 454–462. [DOI] [PubMed] [Google Scholar]
- Bernal M., Casero D., Singh V., Wilson G.T., Grande A., Yang H., Dodani S.C., Pellegrini M., Huijser P., Connolly E.L., Merchant S.S., Krämer U. (2012). Transcriptome sequencing identifies SPL7-regulated copper acquisition genes FRO4/FRO5 and the copper dependence of iron homeostasis in Arabidopsis. Plant Cell 24: 738–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl R.P., Jach G., Saedler H., Huijser P. (2005). Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 352: 585–596. [DOI] [PubMed] [Google Scholar]
- Bowler C., Benvenuto G., Laflamme P., Molino D., Probst A.V., Tariq M., Paszkowski J. (2004). Chromatin techniques for plant cells. Plant J. 39: 776–789. [DOI] [PubMed] [Google Scholar]
- Burkhead J.L., Reynolds K.A., Abdel-Ghany S.E., Cohu C.M., Pilon M. (2009). Copper homeostasis. New Phytol. 182: 799–816. [DOI] [PubMed] [Google Scholar]
- Carey M. (1998). The enhanceosome and transcriptional synergy. Cell 92: 5–8. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S., Ang L.H., Puente P., Deng X.W., Wei N. (1998). Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10: 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Chory J., Fankhauser C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38: 87–117. [DOI] [PubMed] [Google Scholar]
- Chory J., Peto C., Feinbaum R., Pratt L., Ausubel F. (1989). Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 58: 991–999. [DOI] [PubMed] [Google Scholar]
- Cluis C.P., Mouchel C.F., Hardtke C.S. (2004). The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J. 38: 332–347. [DOI] [PubMed] [Google Scholar]
- Cohu C.M., Pilon M. (2007). Regulation of superoxide dismutase expression by copper availability. Physiol. Plant. 129: 747–755. [Google Scholar]
- Das P.K., Geul B., Choi S.B., Yoo S.D., Park Y.I. (2011). Photosynthesis-dependent anthocyanin pigmentation in Arabidopsis. Plant Signal. Behav. 6: 23–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri R.V., Sun H., Palaniswamy S.K., Matthews N., Molina C., Kurtz M., Grotewold E. (2003). AGRIS: Arabidopsis Gene Regulatory Information Server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feild T.S., Lee D.W., Holbrook N.M. (2001). Why leaves turn red in autumn: The role of anthocyanins in senescing leaves of red-osier dogwood. Plant Physiol. 127: 566–574. [PMC free article] [PubMed] [Google Scholar]
- Feng S., Shen Y., Sullivan J.A., Rubio V., Xiong Y., Sun T.P., Deng X.W. (2004). Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein degradation. Plant Cell 16: 1870–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson S.I. (2005). Control of plant development and gene expression by sugar signaling. Curr. Opin. Plant Biol. 8: 93–102. [DOI] [PubMed] [Google Scholar]
- Hobo T., Kowyama Y., Hattori T. (1999). A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. USA 96: 15348–15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes N.M., Neufeld H.S., Burkey K.O. (2005). Functional role of anthocyanins in high-light winter leaves of the evergreen herb Galax urceolata. New Phytol. 168: 575–587. [DOI] [PubMed] [Google Scholar]
- Jiao Y., Lau O.S., Deng X.W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8: 217–230. [DOI] [PubMed] [Google Scholar]
- Joliot P., Joliot A. (2006). Cyclic electron flow in C3 plants. Biochim. Biophys. Acta 1757: 362–368. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14: 787–799. [DOI] [PubMed] [Google Scholar]
- Kirschbaum M.U. (2011). Does enhanced photosynthesis enhance growth? Lessons learned from CO2 enrichment studies. Plant Physiol. 155: 117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat J., Tottey S., Birkenbihl R.P., Depège N., Huijser P., Merchant S. (2005). A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. USA 102: 18730–18735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis S.E., Lilley K.S., Jarvis P. (2008). Isolation and preparation of chloroplasts from Arabidopsis thaliana plants. Methods Mol. Biol. 425: 171–186. [DOI] [PubMed] [Google Scholar]
- Kwok S.F., Staub J.M., Deng X.W. (1999). Characterization of two subunits of Arabidopsis 19S proteasome regulatory complex and its possible interaction with the COP9 complex. J. Mol. Biol. 285: 85–95. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara P., Oñate-Sánchez L., Abraham Z., Ferrándiz C., Díaz I., Carbonero P., Vicente-Carbajosa J. (2003). Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J. Biol. Chem. 278: 21003–21011. [DOI] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.X., Howell S.H. (2010). bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22: 782–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M. (2005). BiNGO: A Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics 21: 3448–3449. [DOI] [PubMed] [Google Scholar]
- Marschner H. (2002). Mineral Nutrition in Higher Plants. (London: Academic Press; ). [Google Scholar]
- Maxwell K., Johnson G.N. (2000). Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 51: 659–668. [DOI] [PubMed] [Google Scholar]
- Merzlyak M.N., Chivkunova O.B., Solovchenko A.E., Naqvi K.R. (2008). Light absorption by anthocyanins in juvenile, stressed, and senescing leaves. J. Exp. Bot. 59: 3903–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol J, Jenkins G., Schafer E., Weiss D. (1996). Signal perception, transduction, and gene expression involved in anthocyanin biosynthesis. Crit. Rev. Plant Sci. 15: 525–557. [Google Scholar]
- Moore D.L., Goldberg J.L. (2011). Multiple transcription factor families regulate axon growth and regeneration. Dev. Neurobiol. 71: 1186–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J.L., Page M.D., Alder N.P., Eriksson M., Quinn J., Soto F., Theg S.M., Hippler M., Merchant S. (2002). Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14: 673–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagae M., Nakata M., Takahashi Y. (2008). Identification of negative cis-acting elements in response to copper in the chloroplastic iron superoxide dismutase gene of the moss Barbula unguiculata. Plant Physiol. 146: 1687–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill S.O., Gould K.S. (2003). Anthocyanins in leaves: Light attenuators or antioxidants? Funct. Plant Biol. 30: 865–873. [DOI] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466. [DOI] [PubMed] [Google Scholar]
- Oyama T., Shimura Y., Okada K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11: 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Lee N., Kim W., Lim S., Choi G. (2011). ABI3 and PIL5 collaboratively activate the expression of SOMNUS by directly binding to its promoter in imbibed Arabidopsis seeds. Plant Cell 23: 1404–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P., Scharfenberg M., Weigel M., Granlund I., Schröder W.P., Finazzi G., Rappaport F., Masiero S., Furini A., Jahns P., Leister D. (2009). Mutants, overexpressors, and interactors of Arabidopsis plastocyanin isoforms: Revised roles of plastocyanin in photosynthetic electron flow and thylakoid redox state. Mol. Plant 2: 236–248. [DOI] [PubMed] [Google Scholar]
- Pilon M., Abdel-Ghany S.E., Cohu C.M., Gogolin K.A., Ye H. (2006). Copper cofactor delivery in plant cells. Curr. Opin. Plant Biol. 9: 256–263. [DOI] [PubMed] [Google Scholar]
- Quinn J.M., Merchant S. (1995). Two copper-responsive elements associated with the Chlamydomonas Cyc6 gene function as targets for transcriptional activators. Plant Cell 7: 623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J.M., Nakamoto S.S., Merchant S. (1999). Induction of coproporphyrinogen oxidase in Chlamydomonas chloroplasts occurs via transcriptional regulation of Cpx1 mediated by copper response elements and increased translation from a copper deficiency-specific form of the transcript. J. Biol. Chem. 274: 14444–14454. [DOI] [PubMed] [Google Scholar]
- Ramshaw J.A., Brown R.H., Scawen M.D., Boulter D. (1973). Higher plant plastocyanin. Biochim. Biophys. Acta 303: 269–273. [DOI] [PubMed] [Google Scholar]
- Schultz T.F., Medina J., Hill A., Quatrano R.S. (1998). 14-3-3 proteins are part of an abscisic acid-VIVIPAROUS1 (VP1) response complex in the Em promoter and interact with VP1 and EmBP1. Plant Cell 10: 837–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T., Müller-Moulé P., Munekage Y., Niyogi K.K., Pilon M. (2003). PAA1, a P-type ATPase of Arabidopsis, functions in copper transport in chloroplasts. Plant Cell 15: 1333–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Park E., Choi G. (2007). PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 49: 981–994. [DOI] [PubMed] [Google Scholar]
- Solfanelli C., Poggi A., Loreti E., Alpi A., Perata P. (2006). Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol. 140: 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F., Kropat J., Malasarn D., Grossoehme N.E., Chen X., Giedroc D.P., Merchant S.S. (2010). The CRR1 nutritional copper sensor in Chlamydomonas contains two distinct metal-responsive domains. Plant Cell 22: 4098–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., et al. (2008). DNA-binding study identifies C-box and hybrid C/G-box or C/A-box motifs as high-affinity binding sites for STF1 and LONG HYPOCOTYL5 proteins. Plant Physiol. 146: 1862–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Kapoor A., Zhu J.K. (2006). Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18: 2051–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F., Habricot Y., Condiff A.S., Maldiney R., Van der Straeten D., Ahmad M. (2007). HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant J. 49: 428–441. [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687. [DOI] [PubMed] [Google Scholar]
- Weigel M., Varotto C., Pesaresi P., Finazzi G., Rappaport F., Salamini F. Leister D. (2003). Plastocyanin is indispensable for photosynthetic electron flow in Arabidopsis thaliana. J. Biol. Chem. 278: 31286–31289. [DOI] [PubMed] [Google Scholar]
- Yadav V., Kundu S., Chattopadhyay D., Negi P., Wei N., Deng X.W., Chattopadhyay S. (2002). Light regulated modulation of Z-box containing promoters by photoreceptors and downstream regulatory components, COP1 and HY5, in Arabidopsis. Plant J. 31: 741–753. [DOI] [PubMed] [Google Scholar]
- Yamasaki H., Abdel-Ghany S.E., Cohu C.M., Kobayashi Y., Shikanai T., Pilon M. (2007). Regulation of copper homeostasis by micro-RNA in Arabidopsis. J. Biol. Chem. 282: 16369–16378. [DOI] [PubMed] [Google Scholar]
- Yamasaki H., Hayashi M., Fukazawa M., Kobayashi Y., Shikanai T. (2009). SQUAMOSA promoter binding protein-like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21: 347–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S. (2000). Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant J. 21: 281–288. [DOI] [PubMed] [Google Scholar]
- Zhang H., Li L. (2013). SQUAMOSA promoter binding protein-like7 regulated microRNA408 is required for vegetative development in Arabidopsis. Plant J. 74: 98–109. [DOI] [PubMed] [Google Scholar]
- Zhang H., He H., Wang X., Wang X., Yang X., Li L., Deng X.W. (2011). Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 65: 346–358. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Zhang H., Li L. (2013). Identification and analysis of the proximal promoters of microRNA genes in Arabidopsis. Genomics 101: 187–194. [DOI] [PubMed] [Google Scholar]
- Zhu X.G., Long S.P., Ort D.R. (2010). Improving photosynthetic efficiency for greater yield. Annu. Rev. Plant Biol. 61: 235–261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.