Figure 4.
SPL7 and HY5 Cobind to the MIR408 Promoter.
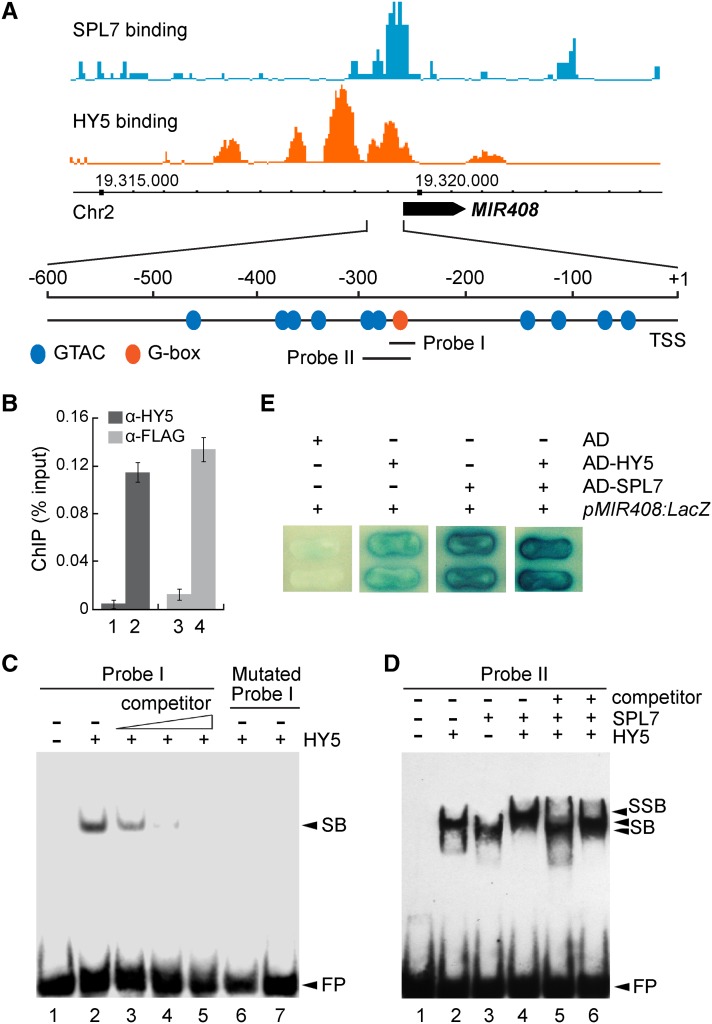
(A) SPL7 and HY5 occupancy at the MIR408 locus based on global ChIP data, which were mapped onto the Arabidopsis genome coordinates and visualized using the Affymetrix Integrated Genome Brower. The position of pre-miR408 is depicted as a black arrow. Orange and blue ovals in the promoter region represent the GTAC and G-box-like motifs, respectively. Probes used in subsequent EMSA analyses are indicated.
(B) Confirmation of HY5 and SPL7 binding to the MIR408 promoter by ChIP-qPCR analysis. ChIP was performed in the indicated genotypes using either the anti-HY5 or anti-FLAG antibody. Data are means ± sd (n = 3). The numbers 1 to 4 denote hy5, the wild type, spl7, and 35S:FLAG-SPL7, respectively.
(C) EMSA analysis of HY5 binding to the G-box in the MIR408 promoter (probe I in A). Lane 1, labeled probe alone; lane 2, labeled probe incubated with recombinant HY5; lanes 3 to 5, excessive unlabeled probe was added as a competitor with the following competitor:probe ratios: 50 (lane 3), 100 (lane 4), and 200 (lane 5); lanes 6 and 7, a mutated probe (CACGTG changed to CTGCAG) incubated with the same amount of HY5 as in lanes 2 to 5 or eight times more HY5, respectively. FP, free probe.
(D) Cobinding of SPL7 and HY5 to the MIR408 promoter revealed by EMSA. Labeled probe II was incubated without protein (lane 1), with HY5 alone (lane 2), SPL7 alone (lane 3), both HY5 and SPL7 (lane 4), HY5 plus SPL7 and 200-fold excessive mutated probe (CACGTG changed to CTGCAG) as a competitor (lane 5), and HY5 plus SPL7 and 200-fold excess mutated probe (GTAC changed to CATG) as a competitor (lane 6). SB, shift band; SSB, super shift band.
(E) Yeast one-hybrid assay testing the cobinding of SPL7 and HY5 to the MIR408 promoter. Yeast cells containing pMIR408:LacZ were transformed with HY5, SPL7, or HY5 plus SPL7 fused with the Gal4 activation domain (AD) and grown on medium containing X-Gal. Cells expressing the activation domain alone were used as the negative control.