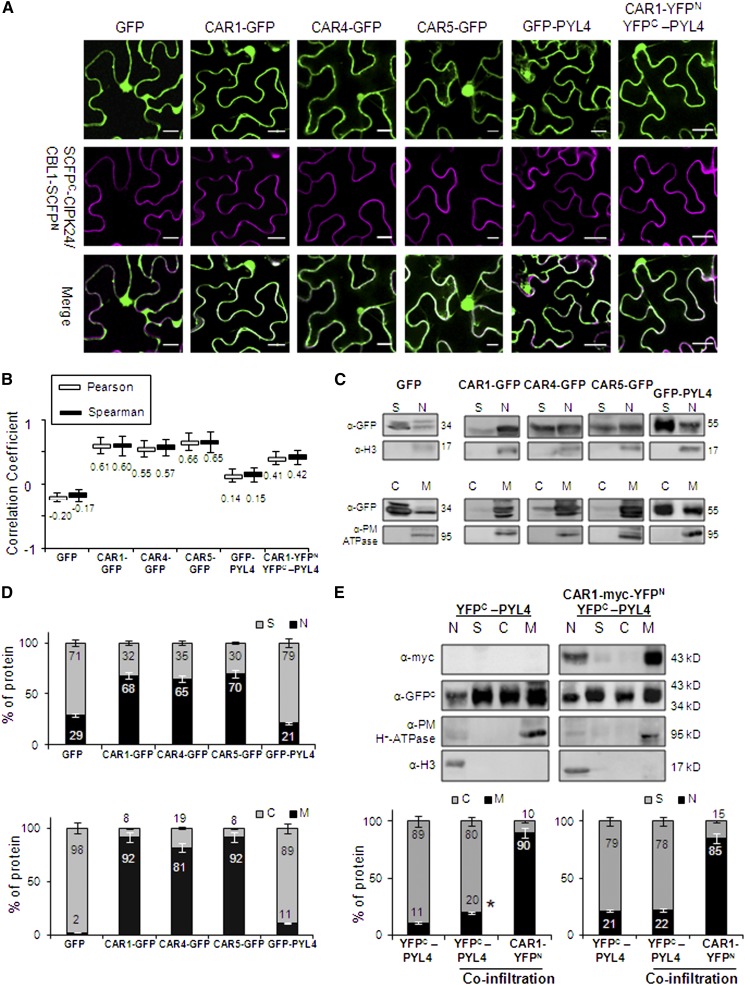
Figure 2.
CAR-GFP Fusion Proteins Localize to Nucleus, Plasma Membrane, and Cytosol upon Transient Expression in N. benthamiana.
(A) Confocal images of transiently transformed N. benthamiana epidermal cells coexpressing GFP, GFP-tagged CAR/PYL4 proteins or the CAR1-YFPN/YFPC-PYL4-interacting proteins, and the plasma membrane marker resulting from the SCFPN-CIPK24/CBL1-SCFPC interaction. The degree of colocalization between the two fluorescent signals was analyzed using merged images and Zeiss software (ZEN Lite 2012). The magenta color of the reconstituted SCFP is a pseudocolor generated from the original cyan fluorescence. Bars = 20 μm.
(B) Pearson-Spearman correlation coefficients indicate the colocalization of CAR-GFP proteins or the CAR1-YFPN/YFPC-PYL4 interaction and the plasma membrane marker. Epifluorescence confocal images of epidermal leaves coinfiltrated with the indicated constructs were merged to quantitatively estimate the colocalization of GFP/YFP and SCFP fluorescence (French et al., 2008). At least 10 single-scanned cell images per experiment were collected and analyzed using the same conditions of laser intensity, pinhole size, and gain levels.
(C) Biochemical fractionation and immunoblot analyses of protein extracts prepared from N. benthamiana leaves infiltrated with Agrobacterium harboring the indicated constructs. Protein extracts from the different fractions were analyzed by immunoblotting using anti-GFP, anti-Histone3 (H3), and anti-plasma membrane (PM) H+-ATPase antibodies. The positions of the molecular mass standards (kD) are indicated. C, cytosolic fraction; M, microsomal fraction; N, nuclear fraction; S, nonnuclear soluble fraction.
(D) Quantification of the subcellular localization of GFP and GFP-tagged proteins transiently expressed in N. benthamiana epidermal cells. Immunoblot signals obtained in (C) were captured using the image analyzer LAS3000, and quantification of the protein signal was done using Image Guache version 4.0 software.
(E) Overexpression of CAR1 together with PYL4 in coinfiltration experiments increases the PYL4 presence in membranes. Biochemical fractionation, immunoblot analyses, and quantification of protein extracts prepared from N. benthamiana leaves infiltrated with Agrobacterium harboring the indicated constructs were performed.