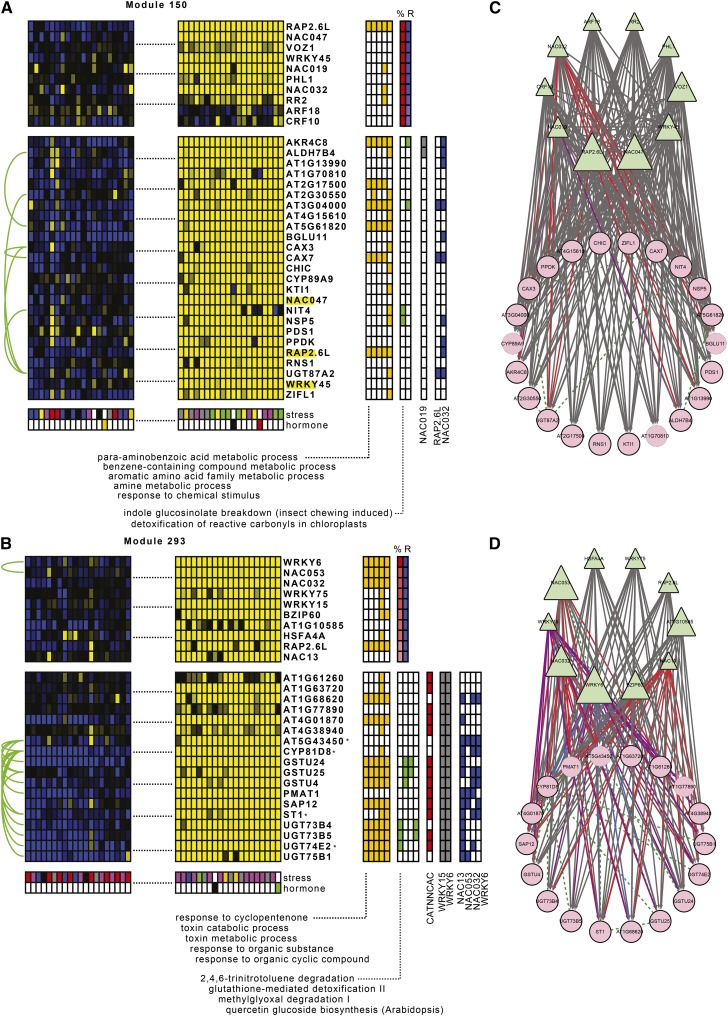
Figure 4.
Experimental and Literature-Based Evidence for Modules Involved in Detoxification Processes of the Oxidative Stress Response.
(A) and (C) Module 150 had as top significant GO Biological Process term “PABA metabolism.” Conditions where the module genes were most highly upregulated consisted of osmotic, salt, and oxidative stress. Two regulatory interactions with NAC019 were confirmed by the extended reference set. Out of 15 module genes tested by nCounter, we experimentally validated 10 targets for NAC032, all of which contained multiple NAC binding motifs in their promoters, and three targets for RAP2.6L. Additionally, we detected one interaction with RAP2.1, four interactions with WRKY6, seven with ERF6, eight with NAC053, and 14 with NAC13.
(B) and (D) Module 293 was highly significantly GO enriched for “response to cyclopentenone” and “toxin catabolic process.” Conditions where the module genes were most highly induced included oxidative stress and auxin inhibition. All module genes were found to be regulated by WRKY6 and WRKY15 in the extended reference set. Through nCounter experiments on 14 module genes, we confirmed four targets for NAC053, seven for WRKY6, eight for NAC032, and 13 for NAC13. Additionally, we found 12 targets for ERF6. Several of these targets contain multiple NAC or W-box binding sites in their promoter, as well as the NAC13/NAC053 MDM motif and the WRKY6 ARE motif. This module is likely involved in MRR.
Interpretation is as in Figure 2. Asterisk indicates MDS gene.