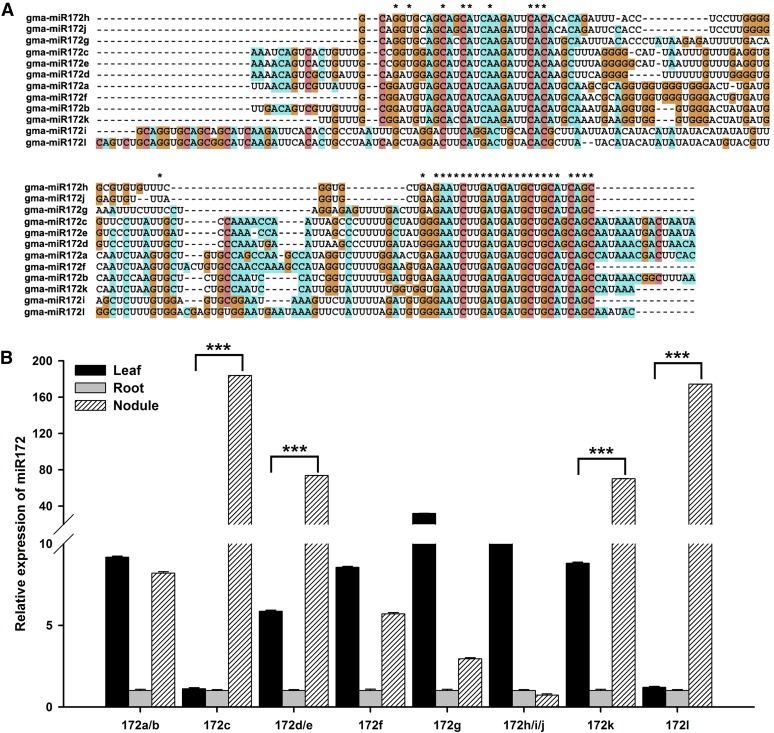
Figure 1.
Sequence Alignments and Tissue-Specific Expression Analysis of miR172 Family Members.
(A) Pre-miRNA sequence alignment of miR172 family members. All sequences of the miR172 family members were aligned using the software MEGA5. Asterisks represent conserved nucleotides in all pre-miRNAs.
(B) Tissue-specific expression analysis of miR172 family members. Seven-day-old seedlings were inoculated with B. japonicum strain USDA110. Leaves, roots, and nodules were harvested at 28 DAI (n = 5). Transcript abundance in the different samples was normalized to that of miR1520d. Expression levels are shown as means ± se from three replicates. Asterisks indicate statistically significant differences (***P < 0.001, Student’s t test).
[See online article for color version of this figure.]