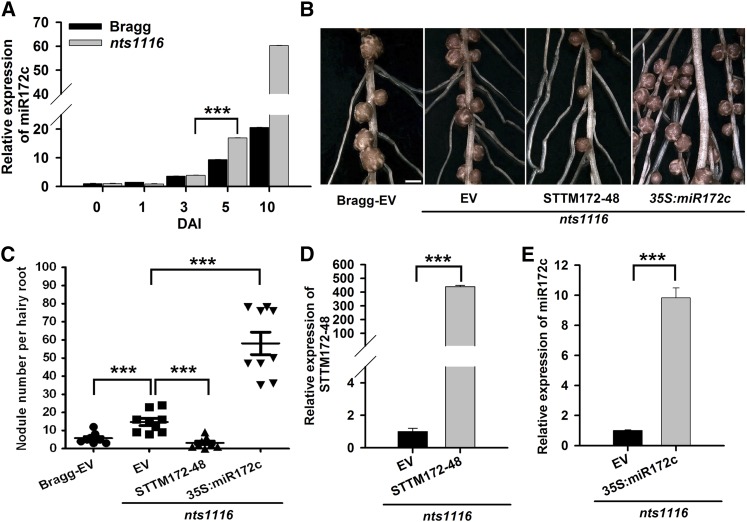
Figure 9.
The Function of miR172c in Regulating Nodule Development Is Negatively Regulated by NARK.
(A) qRT-PCR analysis of miR172c in wild-type cv Bragg and its isogenic nodulation mutant nts1116, which carries a mutation in NARK (n = 10 to 12). miR1520d was used as an internal control for gene expression. Expression levels shown are means ± se from three replicates.
(B) Nodules from hairy roots of nts1116 mutant plants expressing empty vector (EV), STTM172-48, or 35S:miR172c at 28 DAI. Bar = 700 μm.
(C) Quantitative analysis of the nodule number per hairy root of nts1116 mutant plants expressing empty vector, STTM172-48, and 35S:miR172c (n = 10 to 12). Nodule number per hairy root of wild-type cv Bragg plants expressing the empty vector was used as a control. Each value is the average ± sd from three independent experiments. Asterisks represent statistically significant differences (Student’s t test, ***P < 0.001).
(D) qRT-PCR analysis of STTM172-48 in transgenic hairy roots of nts1116 mutant plants (n = 10 to 12). The y axis indicates the expression levels of the gene relative to the expression of ELF1b. Expression levels are means ± se from three replicates. Asterisks represent statistically significant differences (Student’s t test, ***P < 0.001)
(E) qRT-PCR analysis of miR172c in transgenic hairy roots of nts1116 mutant plants. The expression levels were normalized against the geometric mean of miR1520d. Expression levels are means ± se from three replicates. Asterisks represent statistically significant differences (Student’s t test, ***P < 0.001).
[See online article for color version of this figure.]