N-glycosylation of the S-locus receptor kinase ensures its localization to the stigma epidermal cell membrane, where it interacts with its pollen coat-localized ligand to inhibit self-pollination.
Abstract
The S-locus receptor kinase SRK is a highly polymorphic transmembrane kinase of the stigma epidermis. Through allele-specific interaction with its pollen coat-localized ligand, the S-locus cysteine-rich protein SCR, SRK is responsible for recognition and inhibition of self pollen in the self-incompatibility response of the Brassicaceae. The SRK extracellular ligand binding domain contains several potential N-glycosylation sites that exhibit varying degrees of conservation among SRK variants. However, the glycosylation status and functional importance of these sites are currently unclear. We investigated this issue in transgenic Arabidopsis thaliana stigmas that express the Arabidopsis lyrata SRKb variant and exhibit an incompatible response toward SCRb-expressing pollen. Analysis of single- and multiple-glycosylation site mutations of SRKb demonstrated that, although five of six potential N-glycosylation sites in SRKb are glycosylated in stigmas, N-glycosylation is not important for SCRb-dependent activation of SRKb. Rather, N-glycosylation functions primarily to ensure the proper and efficient subcellular trafficking of SRK to the plasma membrane. The study provides insight into the function of a receptor that regulates a critical phase of the plant life cycle and represents a valuable addition to the limited information available on the contribution of N-glycosylation to the subcellular trafficking and function of plant receptor kinases.
INTRODUCTION
In the Brassicaceae, the interaction between pollen grains and the epidermal cells of the stigma surface is regulated by two highly polymorphic proteins: the S-locus receptor kinase (SRK), a single-pass transmembrane kinase that spans the plasma membrane of stigma epidermal cells, and its ligand, the pollen coat-localized S-locus cysteine-rich (SCR) protein. These proteins are encoded by two tightly linked genes of the S locus, variants of which (called S haplotypes) determine specificity in the self-incompatibility (SI) response, an intraspecific mating barrier that promotes outcrossing by preventing self-pollination. SRK and SCR exhibit allele-specific interaction, such that only SRK and SCR variants encoded by the same S haplotype are able to interact (Kachroo et al., 2001; Takayama et al., 2001). Consequently, it is only when stigma epidermal cell and pollen express the same S haplotype (typically in a self-pollination) that SCR binds to the extracellular domain of SRK, thus causing activation of the receptor and the triggering of an SI response that culminates in the inhibition of pollen tube growth at the surface of the stigma epidermis.
Amino acid sequence analysis shows the presence in the extracellular ligand binding domain of SRK of several N-glycosylation motifs that conform to the consensus sequence Asn-X-Ser/Thr (N-X-S/T), in which X can be any amino acid except proline. The presence of these N-glycosylation motifs and previous analysis of soluble stigma-expressed glycoproteins that share a high degree of sequence similarity with the extracellular domain of SRK (Takayama et al., 1989; Umbach et al., 1990) suggest that SRK is glycosylated. However, it is currently not known which of the potential N-glycosylation sites in the SRK extracellular domain are actually glycosylated in planta and if N-glycosylation of the receptor is essential for SI. Although a previous study reported that treating stigmas with the N-glycan modification inhibitor tunicamycin causes breakdown of the SI response (Sarker et al., 1988), it is not known if this breakdown of SI was due to lack of N-glycosylation of the SRK receptor itself or of another protein required for SI.
Analysis of glycosylated receptors in a variety of systems has shown that N-glycans, which are covalently attached to the asparagine residue in N-X-S/T sites, play diverse roles in receptor function, including protection against proteases, correct folding and complex assembly, targeting to the cell surface, and ligand binding (Ohtsubo and Marth, 2006). However, the role of N-glycosylation can vary drastically among different receptors, and this modification is dispensable for the synthesis, processing, or function of some receptors (Everts et al., 1997). Therefore, the potential importance of this modification for the function of a particular receptor must be determined empirically.
In this study, we investigated N-glycosylation of SRK using Arabidopsis thaliana transgenic plants that express the Arabidopsis lyrata SRKb variant. In previous studies, we had shown that SRKb confers intense SI in several accessions of the normally self-fertile A. thaliana, including the C24 accession, and that C24 plants expressing SRKb-mediated SI were a highly suitable platform for analysis of various features of the SRK receptor (Nasrallah et al., 2004; Liu et al., 2007; Boggs et al., 2009b). We therefore used the SRKb variant to assess the functional significance of N-glycosylation of the SRK extracellular domain. We performed a systematic site-directed mutagenesis study and generated mutant versions of SRKb lacking varying numbers of potential N-glycosylation sites. The functionality of mutant SRKb receptors was assessed in transgenic A. thaliana C24 plants by pollinating stigmas expressing these mutant receptors with SCRb-expressing pollen. The accumulation and distribution of wild-type and mutant receptors were analyzed by biochemical and cytological methods. The results show that specific N-glycosylation sites are essential for proper intracellular trafficking of SRK.
RESULTS
The Extracellular Domains of SRK Variants Contain Variable Numbers of Potential N-Glycosylation Sites
We compiled complete or near-complete sequences for the extracellular domains of 12 A. lyrata, 2 Arabidopsis halleri, 38 Brassica oleracea, and 32 Brassica rapa SRKs from public databases (Supplemental Table 1). The number of N-glycosylation motifs in these SRKs ranged from four to nine, and on average, the A. lyrata, A. halleri, B. oleracea, and B. rapa SRKs were found to contain 6.4, 6.0, 7.4, and 7.0 potential N-glycosylation sites, respectively (Supplemental Table 1). The SRK sequences were aligned relative to the sequence of A. lyrata SRKb (accession number BAB40987) using ClustalW (Larkin et al., 2007). Analysis of the sequence alignments (Supplemental Data Set 1) revealed that among the N-glycosylation motifs in SRK variants, three motifs are highly conserved and occur in >90% of SRK variants, while four motifs are moderately conserved and occur in 75 to 86% of SRK variants (Supplemental Data Set 1 and Supplemental Table 2).
The Al-SRK22 variant (accession number ABF71375), which has only four N-glycosylation motifs, the lowest number in all SRK variants analyzed (Supplemental Data Set 1), contains only two of the highly conserved sites (corresponding to amino acid positions 43 and 389 in SRKb) and one of the moderately conserved sites (corresponding to position 260 in SRKb). Moreover, there was some species-specific bias in the conservation of specific N-glycosylation motifs, with some motifs (e.g., those corresponding to positions 115 and 260 in SRKb) being much more highly conserved in Brassica SRKs than in Arabidopsis SRKs. This variability in the number and position of N-glycosylation motifs within and between species suggests that not all potential N-glycosylation sites are critical for SRK structure and function.
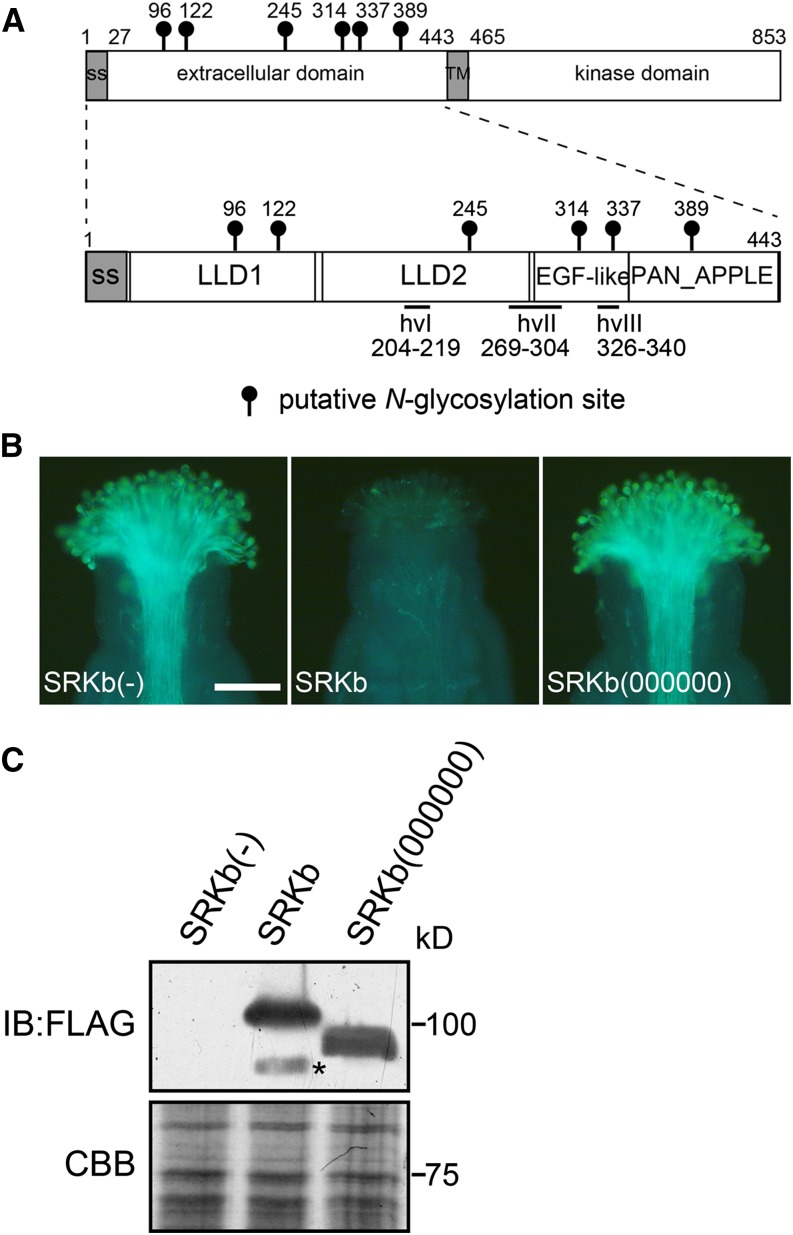
The extracellular domain of SRKb contains six potential N-glycosylation sites: two highly conserved sites (Asn-122 and Asn-389), two moderately conserved sites (Asn-245 and Asn-314), one poorly conserved site (Asn-96), and one site that is unique to this variant (Asn-337) (Figure 1A; Supplemental Table 2). SRKb lacks the most N-terminal highly conserved N-glycosylation motif (amino acid position Lys-43), as well as two moderately conserved motifs (amino acid positions Asp-115 and Asn-260) (Supplemental Table 2). Previous studies had revealed that the SRK extracellular region contains three segments that are hypervariable among SRK variants (hvI, hvII, and hvIII in Figure 1A) (Nishio and Kusaba, 2000; Sainudiin et al., 2005) and are located within a region that determines specificity in the SRK-SCR interaction (Boggs et al., 2009a). Moreover, three-dimensional modeling had shown that this extracellular region consists of four structural subdomains (Figure 1A): two contiguous lectin-like subdomains, designated LLD1 and LLD2, located toward the N terminus; a PAN_APPLE subdomain located at the C terminus; and an intervening region having low structural similarity to epidermal growth factor (EGF) modules, which we refer to as the EGF-like domain (Naithani et al., 2007). As shown in Figure 1A, potential N-glycosylation sites occur in all four structural subdomains of the extracellular domain of SRKb: two in LLD1, one in LLD2, two in the EGF-like domain, and one in the PAN_APPLE domain. Interestingly, only the N337 potential N-glycosylation site, which is unique to SRKb, is located in a hypervariable region of the protein.
Figure 1.
A Requirement of N-Glycosylation Sites in the SRKb Extracellular Domain for the Ability of the Stigma to Inhibit SCRb Pollen.
(A) Schematic structure of the full-length A. lyrata SRKb (top) and its extracellular domain (bottom) showing the location of the four structural subdomains (LLD1, LLD2, EGF-like, and PAN_APPLE) and hypervariable regions (hvI, hvII, and hvIII) that characterize SRK extracellular domains. The positions of the asparagine residues in the N-glycosylation motifs are shown by lollipops.
(B) Microscopic visualization of pollination phenotypes. Representative images are shown for phenotypes observed after pollination with SCRb pollen of stigmas from wild-type untransformed plants lacking SRKb [SRKb(-)] and from transformants expressing wild-type SRKb-FLAG (SRKb) or the mutant SRKb(000000)-FLAG [SRKb(000000)] protein. Note that stigmas expressing wild-type SRKb-FLAG exhibit an intense incompatibility response manifested by the failure of SCRb pollen to germinate and elaborate pollen tubes. By contrast, stigmas expressing the SRKb(000000)-FLAG mutant exhibit a compatible response and allow the growth of numerous SCRb pollen tubes similar to the stigmas of plants lacking SRKb. Bar = 100 μm.
(C) Immunoblot (IB) analysis of SRKb-FLAG proteins from untransformed plants [SRKb(-)] and plants transformed with AtS1pro:SRKb-FLAG [SRKb] and AtS1pro:SRKb(000000)-FLAG [SRKb(000000)]. The upper panel shows immunoblot analysis with anti-FLAG antibody, and the lower panel shows Coomassie blue (CBB) staining as loading control. The asterisk shows a degradation product of SRKb-FLAG.
[See online article for color version of this figure.]
N-Glycosylation of SRKb Is Required for the Ability of the Stigma to Inhibit SCRb-Expressing Pollen
To facilitate analysis of SRK N-glycosylation, we expressed tagged versions of SRKb under the control of the promoter of the At-S1 (Arabidopsis thaliana S-related 1; At3g12000) gene, which is active specifically in stigma epidermal cells (Dwyer et al., 1994). Three tagged versions of the protein, each carrying a different tag, were used: the previously described HA-SRKb, which carries the hemagglutinin (HA) tag at its N terminus (Boggs et al., 2009a); SRKb-cYFP, which carries the citrine variant of yellow fluorescent protein (cYFP) at the C terminus; and SRKb-FLAG, which carries three tandem FLAG tags (3x FLAG) at the C terminus. Similar to HA-SRKb (Boggs et al., 2009a), the SRKb-cYFP and SRKb-FLAG proteins were found to be functional in A. thaliana C24 plants. As illustrated for SRKb-FLAG in Figure 1B (panel SRKb), pollination assays of seven independent AtS1pro:SRKb-FLAG transformants demonstrated that their stigmas inhibited SCRb-expressing pollen (hereafter SCRb pollen), and this inhibition was as intense as that exhibited by the stigmas of C24 plants transformed with untagged SRKb. Thus, neither addition of the 3xFLAG or cYFP tags to the C terminus of full-length SRKb nor addition of the HA tag to its N terminus disrupted receptor function.
To assess the importance of N-glycosylation for SRK function, we generated mutant versions of the AtS1pro:SRKb-FLAG and AtS1pro:HA-SRKb transgenes in each of which all six N-glycosylation motifs of the SRKb extracellular domain were abolished by replacing the serine or threonine residues in the N-X-S/T sites with alanine. These mutant transgenes are hereafter designated AtS1pro:SRKb(000000)-FLAG and AtS1pro:HA-SRKb(000000), where a 0 indicates each of the abolished N-glycosylation sites. Pollination assays in all independent AtS1pro:SRKb(000000)-FLAG transformants (15 plants) and AtS1pro:HA-SRKb(000000) (21 plants) showed that stigmas expressing the mutant proteins failed to inhibit SCRb pollen (Figure 1B, Table 1).
Table 1. Pollination Phenotype of Stigmas Expressing N-Glycosylation Site Mutants of HA-SRKb.
| Transgene | Total No. of Transformants | No. of SCRb Pollen Tubes Produced per Stigma | ||
|---|---|---|---|---|
| <5a | 6–20a | Na | ||
| HA-SRKb | 17 | 14 | 0 | 3 |
| HA-SRKb(000000) | 21 | 0 | 0 | 21 |
| HA-SRKb(011111) | 20 | 18 | 0 | 2 |
| HA-SRKb(101111) | 13 | 1 | 2 | 10 |
| HA-SRKb(110111) | 15 | 12 | 0 | 3 |
| HA-SRKb(111011) | 6 | 6 | 0 | 0 |
| HA-SRKb(111101) | 11 | 9 | 0 | 2 |
| HA-SRKb(111110) | 18 | 0 | 0 | 18 |
| HA-SRKb(001111) | 17 | 0 | 0 | 17 |
| HA-SRKb(100111) | 9 | 0 | 0 | 9 |
| HA-SRKb(101101) | 14 | 0 | 3 | 11 |
| HA-SRKb(010111) | 21 | 3 | 6 | 12 |
| HA-SRKb(011101) | 8 | 4 | 2 | 2 |
| HA-SRKb(110101) | 14 | 11 | 1 | 2 |
| HA-SRKb(010101) | 13 | 0 | 7 | 6 |
For each construct, the table shows the number of transformants in which stigmas exhibited strong, weak, or no incompatibility toward SCRb pollen.
Number of pollen tubes produced per stigma after pollination with SCRb pollen. Intense and weak incompatibility responses are manifested by the growth of <5 and 6 to 20 pollen tubes per stigma, respectively. A compatibility response is manifested by the growth of pollen tubes that were too numerous to count (N).
Consistent with the elimination of all N-glycosylation sites, SRKb(000000)-FLAG and HA-SRKb(000000) exhibited an increased mobility relative to the corresponding tagged versions of wild-type SRKb upon immunoblot analysis of stigma extracts with anti-HA or anti-FLAG antibodies, as illustrated for SRKb(000000)-FLAG in Figure 1C. Importantly, the mutant SRKb proteins accumulated to levels equivalent to those of wild-type SRKb protein (Figure 1C). This result excludes the possibility that suboptimal levels of the SRKb(000000) proteins are responsible for the inability of stigmas to inhibit SCRb pollen. We conclude that lack of N-glycosylation compromises the biogenesis or function of SRKb.
Five of the Six Potential N-Glycosylation Sites in SRKb Are Modified in Planta
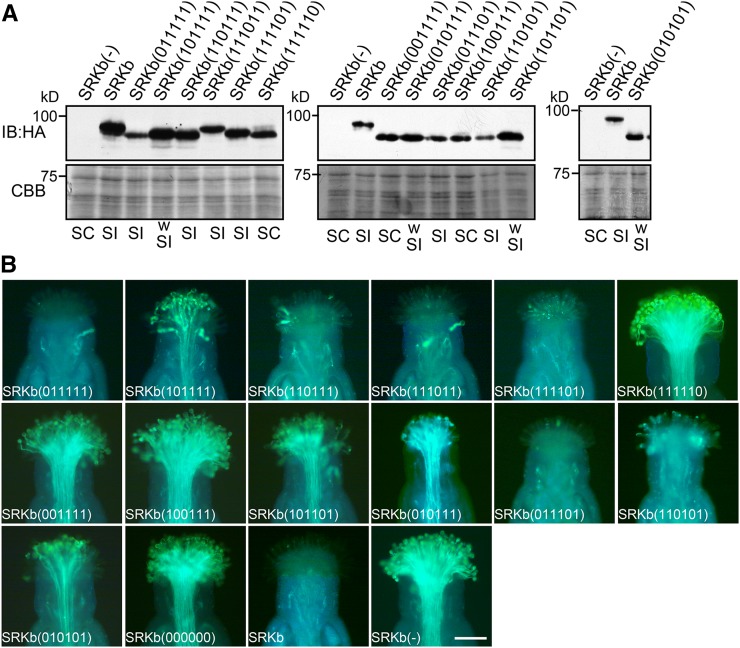
Next, we asked which of the six potential N-glycosylation sites in SRKb was actually modified in stigmas. Toward this end, we used the AtS1pro:HA-SRKb chimeric gene as a template for generating mutant versions of HA-SRKb in each of which one potential N-glycosylation site was abolished by replacing the serine or threonine residue in the N-X-S/T sequence with alanine. The resulting mutant SRKb proteins are designated HA-SRKb(011111), HA-SRKb(101111), HA-SRKb(110111), HA-SRKb(111011), HA-SRKb(111101), and HA-SRKb(111110), in each of which the numbers between parentheses represent the six potential N-glycosylation sites in the order in which they occur in the protein, with 0 indicating the abolished N-glycosylation motif and 1 indicating an intact N-glycosylation motif.
Stigma extracts from transgenic plants expressing each of these mutant SRKb proteins were subjected to immunoblot analysis using anti-HA antibodies. As shown in Figure 2A, and with the exception of HA-SRKb(111011), each of the single N-glycosylation mutants exhibited a slightly increased electrophoretic mobility relative to wild-type HA-SRKb, consistent with the loss of one N-glycan chain. By contrast, the mobility of the HA-SRKb(111011) mutant was identical to that of wild-type HA-SRKb (Figure 2A). These results indicate that the N-glycosylation sites at Asn-96, Asn-122, Asn-245, Asn-337, and Asn-389, but not Asn-314, are actually glycosylated in vivo. The N-glycosylation motif at Asn-314 is N-T-S-P (Supplemental Data Set 1), and its lack of N-glycan modification is consistent with previous reports showing that N-glycosylation is blocked when the amino acid residue at the Y position in the N-X-S/T-Y motif is a proline (Gavel and von Heijne, 1990; Mellquist et al., 1998). Interestingly, in all SRK variants having an N-glycosylation motif at the equivalent position, which comprise 75% of all SRKs analyzed, the Y position is also occupied by a proline (Supplemental Data Set 1). This observation suggests that this particular N-glycosylation site is also unmodified in other SRKs. The Asn-314 site was not considered further in our analysis of SRKb N-glycosylation.
Figure 2.
Varying Effects of Single, Double, and Triple N-Glycosylation Mutants of SRKb on the SI Response.
(A) Immunoblot analysis of HA-tagged SRKb proteins. Total proteins from flower buds of untransformed wild-type plants lacking SRKb [SRKb(-)] and plants expressing wild-type HA-SRKb (SRKb) or the indicated N-glycosylation site HA-SRKb mutants were analyzed by immunoblot (IB) analysis with anti-HA antibody (upper panel). Coomassie blue (CBB) staining is shown as loading control (lower panel). Pollination phenotypes, as determined by pollinating stigmas with SCRb pollen, are shown below each lane (SC, self-compatible; SI, self-incompatible; wSI, weakly self-incompatible).
(B) Microscopic visualization of pollination phenotypes. Representative images are shown for phenotypes observed after pollination with SCRb pollen of stigmas that express mutant HA-SRKb proteins carrying single (top row), double (middle row), and triple (leftmost panel in the bottom row) N-glycosylation site mutations, stigmas expressing HA-SRKb(000000), stigmas expressing wild-type SRKb (SRKb), and stigmas lacking SRKb [SRKb(-)]. Bar = 100 μm.
[See online article for color version of this figure.]
Single, Double, and Triple N-Glycosylation Site Mutants Reveal the Importance of Specific Sites in SRKb for the Ability of the Stigma to Mount the SI Response
Pollination assays of transgenic stigmas with SCRb pollen were performed to assess the effect of eliminating individual N-glycosylation sites on the stigma’s ability to inhibit SCRb pollen. As shown in Table 1, some variability was observed among independent transgenic plants generated by transformation with a particular construct with respect to both pollination phenotype and the number of plants that exhibited a particular phenotype. Such variability is typically observed in plant transformation experiments and may be ascribed to differences in transgene expression levels between transformants. Therefore, for each SRKb mutant construct analyzed in this study, the pollination phenotype of the majority of independent transgenic plants generated, together with the proportion of independent transformants that exhibited this phenotype, was used to assign a phenotype for a mutant SRKb protein with respect to its ability to confer an SI response and the strength of this response.
An incompatible response that was as intense as that observed in stigmas expressing wild-type HA-SRKb was observed in the majority of plants transformed with AtS1pro:HA-SRKb(011111) (18/20 independent transformants), AtS1pro:HA-SRKb(110111) (12/15 independent transformants), and AtS1pro:HA-SRKb(111101) (9/11 independent transformants) (Figure 2B, Table 1). This result indicates that elimination of individual N-glycans at the Asn-96, Asn-245, and Asn-337 residues in SRKb does not disrupt the structure, processing, or function of the protein. By contrast, in all 19 independent AtS1pro:HA-SRKb(111110) transformants analyzed, the stigmas failed to inhibit SCRb pollen (Figure 2B, Table 1). Indeed, the growth of SCRb pollen tubes was as profuse on these stigmas as on the stigmas of AtS1pro:HA-SRKb(000000) transformants or wild-type C24 plants lacking SRKb (Figure 2B). It should be noted that the compatibility of HA-SRKb(111110)-expressing stigmas toward SCRb pollen was not due to suboptimal levels of the mutant SRKb protein because the level of SRKb protein was higher in these stigmas than in stigmas expressing the HA-SRKb(011111) mutant, which confers a robust incompatibility response toward SCRb pollen (Figures 2A and 2B).
As for the AtS1pro:HA-SRKb(101111) construct, in which the N-glycosylation site at Asn-122 was abolished, the majority of the 13 independent transformants analyzed had stigmas that accumulated appreciable amounts of SRKb protein (Figure 2A). Nevertheless, these plants were either fully compatible with SCRb pollen (10 plants) or they exhibited a weak incompatibility response (two plants) toward this pollen (Figure 2B, Table 1). In only one AtS1pro:HA-SRKb(101111) transformant did stigmas exhibit an intense SI response. These results indicate that elimination of N-glycosylation at Asn-122 weakens the ability of the stigma to inhibit SCRb pollen but does not impair the function of SRKb, including its ability to bind to, and become activated by, its SCRb ligand.
We also performed a combinatorial N-glycosylation mutant analysis by constructing AtS1pro:HA-SRKb chimeric genes that carried double and triple mutations of the Asn-96, Asn-122, Asn-245, and Asn-337 N-glycosylation sites. Immunoblot analysis of stigmas expressing each of the resulting HA-SRKb N-glycosylation mutants confirmed that these proteins were expressed at adequate levels (Figure 2A). However, and even though single mutations at the Asn-245 and Asn-337 sites had no effect on the strength of the SI response, the combined mutations caused varying degrees of weakening or loss of SI (Figure 2B, Table 1). Based on the results of this analysis, the four N-glycosylation sites may be arranged in the following order of decreasing importance for SRKb function: Asn-122, Asn-245, Asn-96, and Asn-337. Interestingly, this order parallels the degree of conservation of the four residues among SRK variants (Supplemental Table 2) and is therefore likely to apply to SRKs other than SRKb. This correlation between conservation of an N-glycosylated residue and its importance for the SI response is well illustrated by the Asn-337 site. As stated earlier, this site is unique to SRKb and the only N-glycosylation site that occurs in a hypervariable region of the protein. Polymorphic residues within the hvI, hvII, and hvIII hypervariable regions of the extracellular domain of SRK are generally considered to be responsible for specificity in the SRK-SCR interaction, although empirical evidence for function exists only for the hvI and hvII regions (Boggs et al., 2009a). Only a few SRK variants contain N-glycosylation motifs within their hvIII regions and even fewer within hypervariable regions hvI and hvII (Supplemental Data Set 1). This poor conservation, together with the result of our analysis of the Asn-337 site in SRKb, suggests that N-glycosylation at sites within the hypervariable regions of the SRK extracellular domain is likely to make a relatively minor contribution to the processing or function of SRK.
The N-Glycosylation Motifs in SRKb Are Required for Proper Intracellular Trafficking and Efficient Targeting of the Receptor to the Plasma Membrane
Plasma membrane-localized receptor kinases are transported to the plasma membrane (PM) after folding and assembly in the endoplasmic reticulum (ER) (Benham, 2012). Because N-glycosylation of proteins that are targeted to the cell surface often facilitates proper folding and complex assembly, which are required for exit from the ER and subsequent targeting to the PM (Ruddock and Molinari, 2006), we hypothesized that the SRKb N-glycosylation site mutants that did not confer SI in any of the transformants analyzed might not be targeted efficiently to the PM. To test this hypothesis, three different assays were used to compare the subcellular localization of these mutants to that of wild-type SRKb. First, we used the aqueous two-phase partitioning method, which partitions microsomes into two fractions: one enriched in the PM and another enriched in intracellular membranes (Larsson, 1985; Stein et al., 1996). Microsomes prepared from floral buds collected from AtS1pro:SRKb-FLAG and AtS1pro:SRKb(000000)-FLAG transformants were subjected to two-phase partitioning, and the resulting fractions were used for immunoblot analysis with antibodies raised to the PM-specific marker H+-ATPase, the ER-specific BiP marker, and the FLAG epitope to detect the SRKb proteins. As shown in Figure 3A, the partitioning method achieved substantial enrichment of the PM and intracellular membranes, with only a low level of cross-contamination between the two fractions (Figure 3A).
Figure 3.
Biochemical Analysis of the Effects of N-Glycosylation Site Mutations on the Subcellular Localization of SRKb.
(A) Subcellular distribution of wild-type SRKb-FLAG (SRKb) and mutant SRKb(000000)-FLAG [SRKb(000000)] in fractions enriched for PM and intracellular membranes (non PM). Protein fractions were subjected to immunoblot (IB) analysis with anti-FLAG antibody to detect SRKb-FLAG proteins and with antibodies raised against the H+-ATPase plasma membrane marker or the BiP ER marker to assess specific membrane enrichment.
(B) Sensitivity of SRKb proteins to digestion with Endo H. The left-hand panel shows the results of immunoblot analysis of total proteins extracted from flower buds of wild-type untransformed plants lacking SRKb [SRKb(-)] and transformants expressing wild-type SRKb-FLAG or the SRKb(111110)-FLAG mutant that were left untreated (−) or treated with (+) Endo H at 37°C for 1 h. The upper image shows an immunoblot probed with anti-FLAG antibody and the lower image shows Coomassie blue (CBB) staining as loading control. The right-hand panel shows the results of immunoblot analysis of total proteins extracted from leaf protoplasts that transiently expressed wild-type SRKb-FLAG before and after treatment with Endo H, tunicamycin, or both Endo H and tunicamycin. The glycosylated and deglycosylated forms of SRKb are indicated by arrowheads and arrows, respectively. The asterisks indicate bands that likely represent degradation products of SRKb-FLAG and SRKb(111110)-FLAG. Note that the product of Endo H digestion exhibits the same electrophoretic migration as the deglycosylated SRKb-FLAG proteins produced by cells treated with tunicamycin or with both Endo H and tunicamycin.
The wild-type SRKb-FLAG protein was detected in the PM-enriched fraction, indicating that the protein is localized at the PM. However, it was also detected in the intracellular membrane-enriched fraction (Figure 3A), possibly because high-level expression from the highly active At-S1 promoter allows detection of the protein as it transits through the secretory pathway. In contrast to SRKb-FLAG, the SRKb(000000)-FLAG mutant protein was detected primarily in the intracellular membrane fraction (Figure 3A). The very small amount of SRKb(000000)-FLAG detected in the PM fraction parallels the small amount of the ER marker BiP present in this fraction and is therefore likely due to contamination of the PM fraction with ER and possibly other intracellular membranes. We conclude that SRKb(000000)-FLAG protein is transported very inefficiently, if at all, to the PM.
In a second assay, we compared the subcellular localization of wild-type SRKb-FLAG and the SRKb(111110)-FLAG mutant by examining the electrophoretic migration of these proteins before and after treatment with Endoglycosidase H (Endo H). Endo H specifically cleaves the high mannose-type N-glycans that are attached to proteins in the ER but does not cleave Golgi-modified complex N-glycans (Maley et al., 1989). Consequently, the sensitivity or resistance of N-glycans to Endo H digestion is commonly used as a convenient assay for determining if secreted glycoproteins contain complex N-glycans (i.e., show resistance to Endo H digestion), an indication that they have exited the ER and entered the Golgi apparatus on their way to the cell surface. In plants, sensitivity to Endo H digestion has been used to assess the subcellular localization of the BRASSINOSTEROID INSENSITIVE1 (Jin et al., 2007, 2009; Hong et al., 2008), FLAGELLIN SENSITIVE2 (FLS2) (Lee et al., 2011), and EFR (Nekrasov et al., 2009) receptors and specifically to assay for receptor exit from the ER.
To assay for the sensitivity of wild-type SRKb and the SRKb(111110)-FLAG mutant to Endo H, floral bud extracts were treated with Endo H and analyzed by immunoblotting with anti-FLAG antibody. As shown in Figure 3B, wild-type SRKb-FLAG migrated as a discrete ∼105-kD band in untreated samples, and ∼50% of the molecules in this ∼105-kD band were converted by Endo H to a discrete ∼90-kD band. This result indicates that the ∼105-kD species corresponds to SRKb proteins that contain Endo H-resistant complex N-glycans. For their part, the ∼90-kD SRKb-FLAG species are similar in molecular mass to the 94.1 kD predicted from the amino acid sequence of the SRKb protein, suggesting that they correspond to the deglycosylated form of the protein. This possibility was confirmed by analysis of SRKb-FLAG proteins extracted from cells treated with tunicamycin, an inhibitor of N-glycosylation that causes cells to produce proteins lacking N-glycans. Because tunicamycin treatment cannot be efficiently performed in stigma epidermal cells, we utilized an A. thaliana leaf protoplast expression system in which SRKb-FLAG was expressed transiently (see Methods). As shown in Figure 3B, when treated with Endo H, the deglycosylated SRKb-FLAG protein extracted from tunicamycin-treated cells exhibited the same electrophoretic shift from an ∼105 kD species to an ∼90-kD species as was observed for Endo H-treated SRKb-FLAG from floral bud extracts. Moreover, treatment of tunicamycin-treated cell extracts with Endo H did not cause a shift in the mobility of the deglycosylated SRKb-FLAG protein. These results, together with the fact that the ∼90-kD SRKb-FLAG proteins produced by Endo H treatment migrate as a discrete band (rather than a wide fuzzy band), are consistent with complete removal of N-glycans and indicate that the various N-glycans on the SRKb protein were accessible to the Endo H enzyme in our assays. Similar results were obtained for other plant receptors, which also migrate as discrete bands after Endo H treatment (Jin et al., 2007, 2009; Hong et al., 2008; Nekrasov et al., 2009).
Thus, Endo H digestion revealed that ∼50% of wild-type SRKb-FLAG proteins contain complex N-glycans and therefore must have exited the ER and are likely localized at the PM, consistent with the approximately equal distribution of the SRKb-FLAG protein observed in PM and intracellular membrane fractions obtained by two-phase partitioning. By contrast, the entire pool of the SRKb(111110)-FLAG mutant receptor was converted to an ∼90-kD species upon Endo H treatment, and no ∼105-kD forms of this mutant were detected (Figure 3B), indicating that the bulk of this mutant receptor is not targeted to the PM and is likely trapped in the ER.
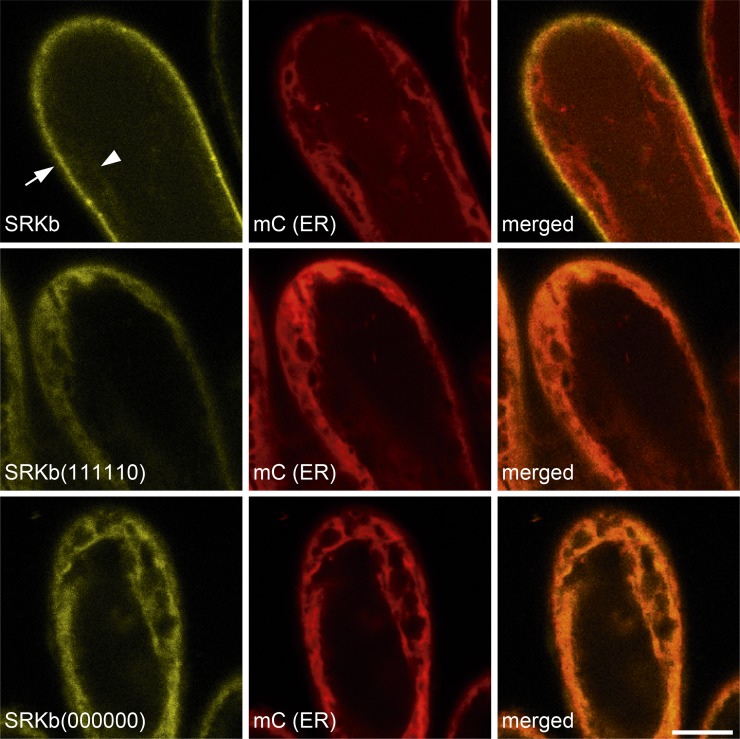
In a third assay, and to determine if the mutant SRKb-cYFP proteins do indeed accumulate in the ER, the localization of cYFP-tagged wild-type and mutant SRKb proteins was visualized by confocal laser scanning microscopy in the stigma epidermal cells of AtS1pro:SRKb-cYFP, AtS1pro:SRKb(000000)-cYFP, and AtS1pro:SRKb(111110)-cYFP transformants that also expressed an ER marker tagged with the mCherry (mC) variant of red fluorescent protein (Nelson et al., 2007; see Methods). Figure 4 shows that wild-type SRKb-cYFP produced an intense signal that was primarily localized at the cell periphery and did not colocalize with the mC-ER marker, consistent with localization of the receptor at the PM. By contrast, the SRKb(000000)-cYFP and SRKb(111110)-cYFP mutant proteins were detected only in intracellular compartments and their signal colocalized perfectly with the mC-ER marker (Figure 4), demonstrating that they accumulate in the ER and are poorly, if at all, targeted to the PM. Together, the biochemical and confocal microscopic data demonstrated that modification of the N389-C390-T391 N-glycosylation site disrupts intracellular trafficking of SRKb and its efficient targeting to the PM, causing accumulation of mutant receptor proteins in the ER.
Figure 4.
Microscopic Analysis of the Effects of N-Glycosylation Site Mutations on the Subcellular Localization of SRKb.
The confocal microscopic images show the results of colocalization experiments of wild-type SRKb-cYFP, SRKb(000000)-cYFP, and SRKb(111110) proteins with an mCherry-labeled ER marker [mC (ER)] in stigma epidermal cells. The images show cYFP (yellow; left column), mC (red; middle column), and merged (right column) images that were captured for the same field of view. Note that wild-type SRKb-cYFP (top row) is primarily localized at the cell periphery (arrow), with only a weak signal observed in intracellular compartments (arrowhead), while the mutant SRKb(000000)-cYFP and SRKb(111110)-cYFP proteins are primarily localized in intracellular compartments (bottom two rows). Also note that the ER marker does not colocalize with the strong wild-type SRKb-cYFP but does colocalize perfectly with SRKb(000000)-cYFP and SRKb(111110)-cYFP proteins. The fact that the strong signals emanating from the ER marker, SRKb(000000)-cYFP, and SRKb(111110)-cYFP are distributed toward the outer edges of the protoplast reflects the presence of a large central vacuole that causes the cytoplasm and ER to be closely appressed to the PM in stigma epidermal cells, as in other plant cells. Bar = 10 μm.
An apparent discrepancy between the results of Endo H treatments and confocal microscopy requires clarification. While Endo H treatment indicated that 50% of wild-type SRKb-cYFP proteins do not exit the ER (Figures 3A and 3B), confocal microscopy of stigmas expressing wild-type SRKb-cYFP failed to detect a strong intracellular cYFP signal. Rather, it showed only a very faint cYFP signal in intracellular compartments, and this signal was only visible upon careful inspection of the confocal images (Figure 4). These apparently conflicting results are most likely due to inherent differences in the surface areas of the ER and the PM. Indeed, similar to the ER in other cells, the ER of stigma epidermal cells has a membrane surface area that is ∼15 to 20 times larger than that of the plasma membrane, as determined by measurements made on transmission electron microscopy images generated in our laboratory (Kandasamy et al., 1989; Umbach et al., 1990), and any SRKb signal that emanates from the ER would be significantly diluted and would not be easily detected by confocal microscopy.
We next asked if the failure or reduced ability of other SRKb N-glycosylation site mutants to confer SI were also correlated with aberrant sensitivity of SRKb to Endo H. To address this issue, we compared the relative proportions of the ∼105- and ∼90-kD SRKb protein species produced upon Endo H treatment of extracts from floral buds that express wild-type SKRb-FLAG to (1) those that express SRKb(001111)-FLAG or SRKb(100111)-FLAG, both of which fail to confer an SI response toward SCRb pollen, and (2) those that express SRKb(101111)-FLAG, which confers a weak SI response (Table 1). As shown in Figure 5, the fractions of the ∼105-kD mutant proteins were drastically reduced upon Endo H treatment of mutant SRKb samples relative to wild-type SRKb-FLAG samples. These reductions were particularly severe in the case of SRKb(001111)-FLAG and SRKb(100111)-FLAG and somewhat less severe in the case of SRKb(101111)-FLAG (Figure 5), consistent with the fact that stigmas expressing these receptors exhibited complete and partial loss of SI, respectively. These results indicate that abolishing the Asn-96, Asn-122, and Asn-245 N-glycosylation sites severely disrupts the proper subcellular trafficking of SRKb proteins. Furthermore, the similarity of the electrophoretic shifts produced upon Endo H treatment of these mutant receptors to those observed for the SRKb(000000)-cYFP and SRKb(111110)-cYFP mutants suggests that the SRKb(001111)-FLAG, SRKb(100111)-FLAG, and SRKb(101111)-FLAG proteins are not, or only very inefficiently, translocated to the PM.
Figure 5.
Sensitivity to Endo H of Additional SRKb N-Glycosylation Mutants That Fail to Confer SI.
Total proteins from flower buds of wild-type untransformed plants lacking SRKb [SRKb(-)], plants expressing wild-type SRKb-FLAG (SRKb), and plants expressing the SRKb(101111), SRKb(001111)-FLAG, and SRKb(100111) mutant proteins were left untreated (−) or treated with (+) Endo H. The glycosylated and deglycosylated forms of SRKb proteins are indicated by arrowheads and arrows, respectively. The asterisks indicate bands that likely represent degradation products of SRKb proteins. Note that the apparent slower mobility of bands in the right-hand lanes relative to those in the other lanes is due to irregular electrophoretic migration of proteins across the gel, as demonstrated by the reduced mobility of the Endo H band within the same lanes (Coomassie blue [CBB] panel).
The Thr-391 Residue in SRKb, but Not N-Glycosylation at Asn-389, Is Required for the SRKb-Mediated SI Response
The N389-C390-T391 N-glycosylation motif is conserved in the vast majority of SRKs but is absent in three SRK variants, namely, B. rapa Br-SRK53 (Supplemental Data Set 1) and the A. halleri variants Ah-SRK2 and Ah-SRK14 (Supplemental Data Set 2), in which the NCT motif is replaced by KCT, NCL, and SCT, respectively. This observation indicates that N-glycosylation at the Asn-389 site is dispensable for SRK function and suggests that the inability of the SRKb(111110) mutant to confer SI might be due to a structural change resulting from the T391A mutation rather than to loss of the N-glycan at the Asn-389 position.
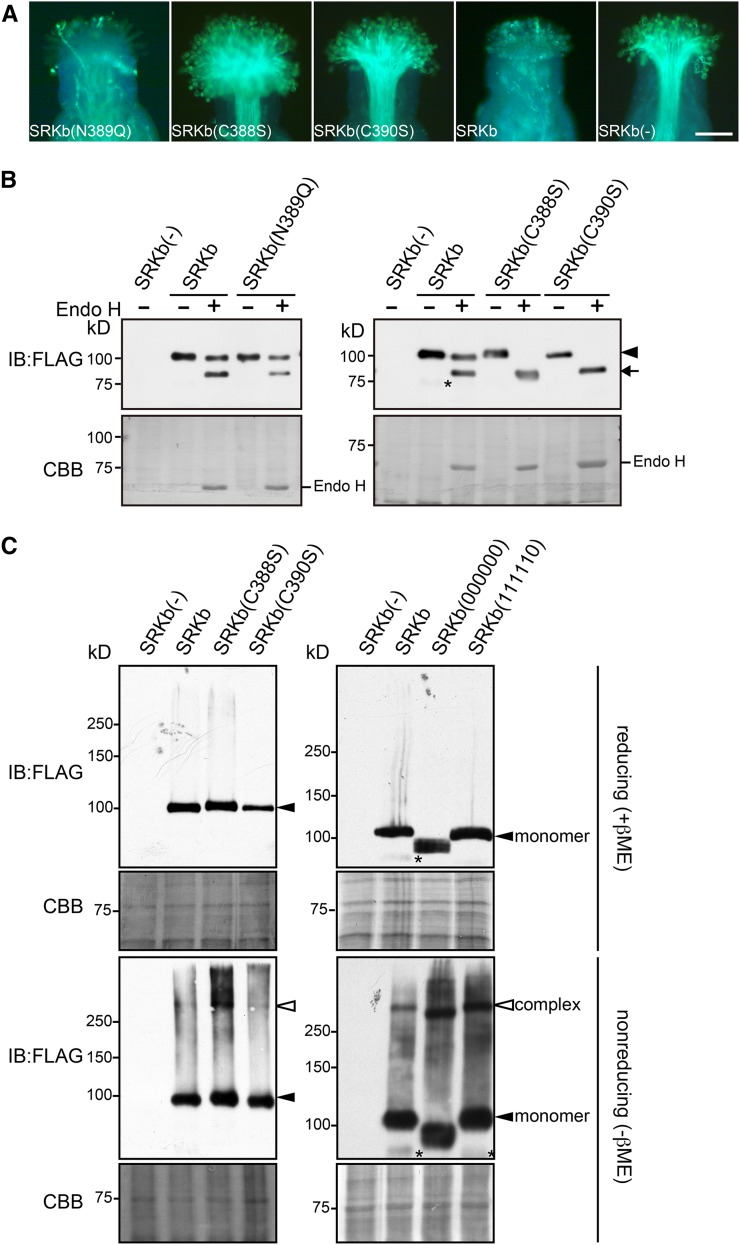
To test this possibility, we generated an SRKb-FLAG mutant in which N-glycosylation at the Asn-389 site was abolished by replacing the Asn-389 residue with glutamine (Q), and we examined the ability of the resulting SRKb(N389Q)-FLAG mutant to confer SI. Unlike AtS1pro:SRKb(111110) transformants, none of which exhibited an incompatible response toward SCRb pollen (Table 1), the stigmas of 10 of 14 AtS1pro:SRKb(N389Q)-FLAG transformants showed an intense incompatible response toward SCRb pollen (Figure 6A, Table 2). Consistent with this phenotype, SRKb(N389Q)-FLAG exhibited the same sensitivity to Endo H as wild-type SRKb-FLAG, i.e., it produced approximately equal proportions of the ∼105- and ∼90-kD protein species upon Endo H treatment (Figure 6B). This result indicates that the Thr-391 residue itself, rather than N-glycosylation at the Asn-389 residue, is critical for exit of SRKb from the ER.
Figure 6.
Functional and Electrophoretic Analysis of SRKb Mutants Carrying Cysteine and N-Glycosylation Motif Mutations in an Essential Region of the SRKb PAN_APPLE Domain.
(A) Microscopic visualization of pollination phenotypes. Representative images are shown for pollination phenotypes observed after pollination with SCRb pollen of stigmas expressing the FLAG-tagged SRKb(N389Q), SRKb(C388S), and SRKb(C390S) mutants, or wild-type SRKb (SRKb), and stigmas from untransformed plants lacking SRKb [SRKb(-)]. Bar = 100 μm.
(B) Sensitivity to Endo H of FLAG-tagged wild-type SRKb, SRKb(N389Q), SRKb(C388S), and SRKb(C390S) proteins. The glycosylated and deglycosylated forms of SRKb proteins are indicated by an arrowhead and arrow, respectively. Sample preparation, enzymatic treatment, and labeling of figure panels were as in Figure 3B. The asterisk indicates a band that likely represents a degradation product of SRKb-FLAG. Note that the SRKb(C388S) and SRKb(C390S) proteins migrate slightly more slowly than wild-type SRKb, likely due to the C-to-S substitution, which has been shown to cause a slight reduction in electrophoretic mobility in other proteins (Sakoh-Nakatogawa et al., 2009).
(C) Detection of disulfide bond-mediated SRKb complexes in stigmas expressing FLAG-tagged wild-type SRKb, SRKb(C388S), SRKb(C390S), SRKb(000000), and SRKb(111110). Total proteins were prepared from flower buds in the presence of 15 mM iodoacetamide, subjected to SDS-PAGE in the presence of β-mercaptoethanol (reducing [+βME]) or in the absence of β-mercaptoethanol (nonreducing [-βME]), followed by immunoblot (IB) analysis with anti-FLAG antibody. Coomassie blue (CBB) staining is shown as loading control. Bands corresponding to SRKb monomers and complexes are indicated. The asterisks show a degradation product of SRKb-FLAG and SRKb(111110).
[See online article for color version of this figure.]
Table 2. Pollination Phenotype of Stigmas Expressing PAN_APPLE Domain Mutants of SRKb-FLAG.
| Transgene | Total No. of Transformants | No. of SCRb Pollen Tubes Produced per Stigma |
||
|---|---|---|---|---|
| <5a | 6–20a | Na | ||
| SRKb-FLAG | 7 | 7 | 0 | 0 |
| SRKb(N389Q)-FLAG | 14 | 10 | 3 | 1 |
| SRKb(C388S)-FLAG | 19 | 0 | 0 | 19 |
| SRKb(C390S)-FLAG | 13 | 0 | 0 | 13 |
Number of pollen tubes produced per pollinated stigma after pollination with SCRb pollen. Pollen tube counts are as in Table 1.
The Thr-391 Residue Adjoins Two Critical Cysteine Residues of the SRK PAN_APPLE Domain
The Thr-391 residue of SRKb is located within the PAN_APPLE domain, which forms the C terminus of the extracellular region of the receptor (Figure 1A). Consistent with the fact that PAN_APPLE domains are typically protein-protein interaction domains (named for their apple-like shape), the PAN_APPLE domain of SRK was previously shown by interaction studies in yeast to be required for ligand-independent dimer formation of the extracellular regions of SRKs (Naithani et al., 2007). Three-dimensional modeling of the SRK PAN_APPLE domain predicts a β-sheet structure consisting of five antiparallel β-sheets (designated β1 through β5) and an α-helix connected to the core β-sheet structure by two disulfide bonds that link four of the six cysteine residues found in this domain (Naithani et al., 2007).
Of particular interest for our study is the loop region between the α-helix and the β3 sheet (Naithani et al., 2007). In both the B. oleracea SRK6 variant originally used for three-dimensional modeling (Naithani et al., 2007) and in the SRKb variant analyzed here, this loop region contains the C388-N389-C390-T391 sequence. In this critical sequence, the Cys-388 residue is predicted to be free and available for disulfide bond formation with cysteine residues outside the PAN_APPLE domain, the Cys-390 residue is predicted to form an intramolecular disulfide bridge with Cys-384, and the Thr-391 residue is essential for PM targeting of SRKb as shown in this study. The Thr-391 residue, which is located at one edge of the β3 sheet, is conserved in all SRK variants except for Ah-SRK2, in which it is replaced by leucine (Supplemental Data Set 2). While leucine, like threonine, is often found in β-sheet structures, the alanine residue introduced by the T391A mutation is infrequently observed in β-sheet regions (Chou and Fasman, 1978). Based on these observations, we hypothesized that the T391A mutation we introduced in SRKb is structurally disruptive and might affect the formation of C390-mediated intramolecular and/or C388-mediated intermolecular disulfide bonds.
We examined the importance of the Cys-388 and Cys-390 residues by replacing each of these cysteines with serine (S) in the AtS1pro:SRKb-FLAG gene. Pollination assays revealed that the stigmas of all plants expressing the resulting SRKb(C388S)-FLAG (19 plants) and SRKb(C390S)-FLAG proteins (13 plants) failed to inhibit SCRb pollen (Figure 6A, Table 2). Thus, the Cys-388 and Cys-390 residues are essential for the SRKb-mediated SI response. Interestingly, both SRKb(C388S)-FLAG and SRKb(C390S)-FLAG proteins exhibited complete sensitivity to Endo H digestion. The lack of complex N-glycan-containing forms of these SRKb mutants (Figure 6B), which is indicative of retention in the ER, suggests that these mutant proteins are unlikely to be transported efficiently, if at all, to the PM.
Proper Disulfide Bridge-Mediated Complex Formation of SRKb Is Required for Targeting of the Receptor to the Plasma Membrane
A possible explanation for the observation that several of the SRKb mutants analyzed in this study exhibit defective intracellular translocation is that these proteins might form aberrant higher-order complexes that cause them to be trapped in intracellular compartments, as shown for several mammalian proteins such as the null Hong Kong variant of human a1-trypsin (Hosokawa et al., 2006) and the unassembled J chains of polymeric immunoglobulins (Mezghrani et al., 2001). In a preliminary test of this possibility, we compared the propensity of wild-type and mutant SRKb-FLAG proteins to form disulfide bridge-mediated dimers and higher-order oligomers. We prepared total proteins from floral buds expressing wild-type SRKb-FLAG, SRKb(C388S)-FLAG, SRKb(C390S)-FLAG, SRKb(000000)-FLAG, and SRKb(111110)-FLAG in the presence of iodoacetamide, a reagent that alkylates thiol groups and prevents spurious disulfide-bridge formation during sample preparation (Hollecker, 1997). The protein samples were subjected to SDS-PAGE under nonreducing and reducing conditions followed by immunoblot analysis with anti-FLAG antibody.
As shown in Figure 6C, only protein species that migrated with the apparent molecular masses expected for the monomeric forms of wild-type and mutant SRKb-FLAG proteins were detected under reducing conditions. By contrast, nonreducing conditions produced complex electrophoretic patterns. In wild-type SRKb-FLAG samples, we observed SRKb-FLAG monomers as well as a higher molecular mass species (Figure 6C) that must correspond to SRKb-FLAG complexes produced by the formation of intermolecular disulfide bridges because they are not detected in the presence of the reducing agent β-mercaptoethanol. A similar electrophoretic pattern consisting of a monomeric and a high molecular mass species was also observed under nonreducing conditions for the SRKb(C388S)-FLAG and SRKb(C390S)-FLAG mutant proteins (Figure 6C), suggesting that cysteine residues other than Cys-388 and Cys-390 contribute to the formation of these higher-order SRKb complexes.
Careful inspection of the electrophoretic patterns generated for wild-type SRKb-FLAG samples under nonreducing conditions showed a faint smear of very slow-migrating molecular species that likely consists of heterogeneous disulfide-bonded aggregates of SRKb with itself or with other molecules (Figure 6C). Interestingly, comparison of wild-type and mutant SRKb-FLAG samples showed that both the amount and molecular mass of these aggregates were increased drastically in SRKb(C388S)-FLAG, SRKb(000000)-FLAG, and SRKb(111110)-FLAG samples, but only moderately in SRKb(C390S)-FLAG samples (Figure 6C). The results suggest that the C388S, C390S, and T391A mutations affect the formation or accumulation of disulfide-mediated SRKb complexes and that the generation of high molecular mass SRKb aggregates is mediated in part by the Cys-390 residue.
DISCUSSION
In this study, we investigated the importance of N-glycosylation for the ability of the SRK receptor to confer SI in transgenic A. thaliana. Our mutagenic analysis of N-glycosylation motifs in the SRKb receptor demonstrated that five of six N-glycosylation motifs found in the SRKb extracellular domain are modified in planta and that abolishing all of these motifs resulted in SRKb molecules that failed to confer SI. However, further analysis of SRKb N-glycosylation site mutants indicated that elimination of N-glycosylation sites either singly or in various double and triple combinations does not disrupt the SCRb-dependent activation of the receptor. Indeed, all but one of the single N-glycosylation mutants and several of the double mutants we analyzed retained the ability to confer an intense SI response, while other double and triple mutants conferred a weakened but still evident SI response. Moreover, even in the case of the one N-glycosylation site, Asn-389 in the N389-C390-T391A motif, whose elimination did abrogate the SI response, our results indicated that it was the structural disruption caused by the T391A mutation rather than the lack of an N-glycan chain at this site that caused breakdown of SI. Thus, SRKb is more similar to the A. thaliana FLS2 (Sun et al., 2012) and the Medicago truncatula NFP (Lefebvre et al., 2012) receptor-like kinases, which are insensitive to disruption of its N-glycosylation sites, than to the A. thaliana EFR and tomato (Solanum lycopersicum) Cf-9 receptors, which are inactivated by disruption of even single N-glycosylation sites (van der Hoorn et al., 2005; Häweker et al., 2010; Saijo, 2010).
Our analysis of SRKb N-glycosylation mutants that fail to confer SI indicates that N-glycosylation of the SRKb extracellular domain functions primarily to ensure the proper and efficient trafficking of SRKb to the PM. Indeed, we found that SRKb N-glycosylation mutant proteins were not detected in the PM fraction upon two-phase partitioning of stigma extracts, that these proteins lacked Endo H-resistant complex N-glycans or contained drastically reduced levels of these N-glycans, and that they colocalized with an ER marker in optical sections of stigma epidermal cells. These observations are all consistent with the conclusion that the SRKb N-glycosylation site mutants are trapped in intracellular compartments, specifically the ER, in contrast to wild-type SRKb, a major fraction of which exits the ER and is targeted to the PM. Thus, the breakdown or weakening of SI observed in stigmas expressing these SRKb N-glycosylation site mutants can be ascribed largely to the failure, or drastically reduced ability, of these proteins to localize to the PM, where they are available for interaction with their pollen-derived SCRb ligand. A similar role for N-glycosylation in the trafficking of plasma membrane-localized proteins was reported in mammalian systems for the EGF receptor (Soderquist and Carpenter, 1984; Slieker et al., 1986) and the Na, K-ATPase β2 subunit (Tokhtaeva et al., 2010).
The mechanism by which elimination of specific N-glycosylation sites disrupts the subcellular trafficking of SRKb remains to be determined. A possibility suggested by the data is that abolishing these N-glycosylation sites might cause the increased formation of disulfide bond-mediated receptor complexes and aggregates. In support of this conclusion, stigmas expressing functionally defective SRKb N-glycosylation mutants or SRKb mutants generated by elimination of essential cysteine residues in the PAN_APPLE domain, which functions in ligand-independent dimer formation of the extracellular regions of SRKs (Naithani et al., 2007), exhibited similar increases in the amount of high molecular mass SRKb aggregates relative to the wild type, and these increases correlated with the lack of complex N-glycan-containing species for these proteins. The composition of these high molecular mass aggregates remains to be determined, but their increase in stigmas expressing mutant SRKb proteins that fail to confer SI suggests that they represent, at least in part, nonfunctional SRKb aggregates. We speculate that the mutations we introduced into this motif affect disulfide-bond formation and that increased aggregation of the mutant SRKb proteins disrupts their export from the ER and their transport to the PM. Further analysis is required to test this hypothesis.
In summary, our results underscore the importance of N-glycosylation at specific residues within the SRKb extracellular domain, of the C388-N389-C390-T391 motif within the PAN_APPLE subdomain and of the structural integrity of this subdomain for proper intracellular trafficking of SRKb and consequently for robust execution of the SI response at the stigma epidermal cell surface. While our investigation of the role of N-glycosylation focused on the SRKb variant, it is important to note that we observed a strong correlation between the degree to which a particular N-glycosylation site was conserved among SRK variants and the extent to which elimination of the site disrupted intracellular translocation of the SRKb receptor. This correlation suggests that the inferences made herein are not specific to SRKb but are likely to apply to other SRK variants as well.
METHODS
Molecular Phylogenetic Analyses
Amino acid sequences were aligned using ClustalW (Larkin et al., 2007) with a Gonnet matrix.
Generation of N-Glycosylation Mutant Transgenes, Plant Transformation, and Plant Growth
All transgenes were inserted into the pCAMBIA1300 plant transformation plasmid (GenBank accession number AF234296). Wild-type and mutant forms of SRKb were expressed from a 360-bp fragment corresponding to the promoter of the At-S1 (At3g12000) gene, which is a highly active stigma epidermal cell-specific promoter (Dwyer et al., 1994; Boggs et al., 2009a). Construction of the AtS1pro:HA-SRKb transgene was described previously (Boggs et al., 2009a). For expression of SRKb proteins that were C-terminally tagged with a 3xFLAG epitope, a sequence encoding three tandem FLAG tags was introduced just before the stop codon of the AtS1pro:SRKb gene by the recombinant PCR method. Similarly, for expression of SRKb carrying a C-terminal fluorescent protein tag, a sequence encoding cYFP preceded by a linker sequence was amplified and inserted by recombinant PCR just before the stop codon of the AtS1pro:SRKb gene. The primers used for transgene construction are listed in Supplemental Table 3.
To introduce N-glycosylation site mutations, the above plasmids served as templates for recombinant PCR-mediated site-directed mutagenesis using mutagenic primers (Supplemental Table 3). All plasmids were sequenced at the Cornell University Life Sciences Core Laboratories Center to exclude the presence of PCR-generated errors. The plasmids were introduced into Agrobacterium tumefaciens strain GV3101 (Koncz and Schell, 1986) and subsequently into Arabidopsis thaliana plants of the C24 accession by the floral dip method (Clough and Bent, 1998). Transformants were selected on Murashige and Skoog medium (Sigma-Aldrich) containing 50 μg/mL hygromycin. Plants were grown in a growth chamber at 22°C under continuous light.
Pollination Assays
To examine the effects of the site-directed mutations introduced in SRKb on the SI response, stigmas expressing each mutant receptor were pollinated with SCRb-expressing pollen grains derived from plants transformed with the Arabidopsis lyrata SRKb-SCRb gene pair (Nasrallah et al., 2004). Stigmas of buds at stage 13 of development (Smyth et al., 1990) were manually pollinated with pollen grains obtained from mature postanthesis flowers under a stereomicroscope. Two hours after pollination, the stigmas were fixed, stained with decolorized aniline blue, and examined by epifluorescence microscopy as previously described (Kho and Baër, 1968). In these assays, an incompatible response of a stigma is manifested by the growth of fewer than five pollen tubes, a partially incompatible response by the growth of 6 to 20 pollen tubes, and a compatible response by the growth of pollen tubes that are too numerous to count. Images of pollinated stigmas were captured using a VANOX-T microscope (Olympus) with a Polaroid DMC camera.
Protein Immunoblot Analysis
Total proteins were extracted from 10 stage 13 flower buds. The flower buds were homogenized in a solution containing 100 mM Tris-HCl, pH 8.0, 2% (w/v) SDS, 5.7 mM β-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride, and the extracted total proteins were precipitated using the trichloroacetic acid/acetone method and dissolved in SDS-PAGE sample buffer (Laemmli, 1970). For nonreducing SDS-PAGE analysis, the flower buds were homogenized in a solution containing 100 mM Tris-HCl, pH 8.0, 2% (w/v) SDS, 15 mM iodoacetamide, and 1 mM phenylmethylsulfonyl fluoride, and the precipitated proteins were dissolved in SDS-PAGE sample buffer lacking β-mercaptoethanol. Samples were subjected to SDS-PAGE on 6% (w/v) gels followed by transfer to Immobilon-P membranes (Millipore) as described by Towbin et al. (1979). For detection of tagged proteins, monoclonal anti-HA antibody (Covance) and anti-FLAG antibody (Sigma-Aldrich) were used at a 1:1000 dilution, and goat anti-mouse peroxidase-labeled antibody (Sigma-Aldrich) was used as secondary antibody at a 1:3000 dilution. The anti-H+-ATPase antibodies (Agrisera) and anti-AtBiP antibodies (Yamamoto et al., 2008) were used at a 1:3000 dilution, and anti-rabbit peroxidase-labeled antibody (CLONE RG-96; Sigma-Aldrich) was used as secondary antibody at a 1:3000 dilution. Immunodetection was performed using the ECL2 system (Thermo Fisher Scientific) and exposure to x-ray film. To confirm equal protein loading for different samples, the protein blots were subsequently stained with Coomassie Brilliant Blue R 250 (Bio-Rad).
Aqueous Two-Phase Partitioning and Endo H Digestion
Isolation of microsomal membranes and aqueous two-phase partitioning were performed according to methods described previously (Larsson, 1985; Stein et al., 1996) as follows. Fifty stage 13 flower buds were homogenized in a solution containing 30 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% (v/v) glycerol, and 5 mM MgCl2. The homogenate was centrifuged at 8200g for 5 min and filtered through a micro bio-spin column (Bio-Rad) to remove cell debris, and the resulting filtrate was centrifuged at 100,000g for 1 h to collect microsomal membranes. The microsomal membranes were resuspended in a buffer consisting of 5 mM potassium phosphate, pH 7.8, 3 mM KCl, and 0.33 M sucrose, and aqueous two-phase partitioning was performed in a series of 1-mL volume systems consisting of 6.2% (w/w) Dextran T500, 6.2% (w/w) polyethylene glycol 3350, 5 mM potassium phosphate buffer, pH 7.8, 3 mM KCl, and 0.33 M sucrose. Three rounds of partitioning yielded an upper phase enriched in plasma membranes and a lower phase enriched in intracellular membranes. Proteins in each fraction were precipitated using the trichloroacetic/acetone method and analyzed by SDS-PAGE and immunoblotting as described above.
For digestion with Endo H (New England Biolabs), total proteins prepared from 10 stage 13 flower buds were denatured and treated with the enzyme at 37°C for 1 h according to the manufacturer’s protocol. The digested samples were analyzed by SDS-PAGE and immunoblotting as described above.
Transient Expression of SRKb-FLAG in A. thaliana Leaf Protoplast Cells and Tunicamycin Treatment
For transient expression of SRKb-FLAG in A. thaliana leaf protoplasts, we used a pUC19 plasmid into which an EcoRI-XbaI fragment containing the 35Spro:SRKb-FLAG:SRKbterm chimeric gene was inserted. The isolation of A. thaliana leaf protoplasts and transient expression methods were performed essentially as described by Yoo et al. (2007). In brief, 50 leaves of 3- or 4-week-old A. thaliana C24 plants were cut into 0.5- to 1-mm leaf sections and incubated for 6 h in 10 mL of enzyme solution (1.5% [w/v] cellulase Onozuka R-10 [Yakult Pharmaceutical], 0.4% [w/v] macerozyme R-10 [Yakult Pharmaceutical], 0.4 M mannitol, 20 mM KCl, and 20 mM MES, pH 5.7). The prepared leaf protoplasts were incubated overnight at room temperature with 5 µg of plasmid DNA in a solution containing 40% (w/v) polyethylene glycol 4000 (Sigma-Aldrich), 0.2 M mannitol, and 100 mM CaCl2. The transfected cells were either left untreated or treated with 5 µg/mL tunicamycin (Wako) for 8 h at room temperature. Extraction of total proteins, precipitation of proteins with trichloroacetic acid/acetone, Endo H treatment, SDS-PAGE, and immunoblotting were as described above.
Confocal Laser Scanning Microscopy
For colocalization studies, an ER marker tagged with the mC variant of RFP (Nelson et al., 2007) was used. A plasmid containing a chimeric gene in which the cauliflower mosaic virus double 35S promoter drives expression of the mC-tagged ER marker (available from the ABRC) was modified by replacing the double cauliflower mosaic virus 35S promoter, which is not highly active in stigma epidermal cells, with the At-S1 promoter using recombinant PCR. A transgenic line homozygous for the resulting AtS1pro:mC-ER transgene was crossed to transgenic lines homozygous for the AtS1pro:SRKb-cYFP, AtS1pro:SRKb(000000)-cYFP, or AtS1pr:SRKb(111110)-cYFP transgenes to generate F1 plants that express both cYFP-tagged SRKb protein and the mC-tagged ER marker. Stigmas from stage 12 flower buds were observed with a Leica TCS-SP5 confocal microscope (Leica Microsystems) using a ×63 water immersion objective (numerical aperture 1.2), zoom 5.0, at the Plant Cell Imaging Facility of the Boyce Thompson Institute (Ithaca, NY). cYFP was excited with the argon laser (15% 514 nm), and emitted fluorescence was collected from 522 to 550 nm; mCherry was excited with the diode-pumped solid state laser (5% 561 nm), and emitted fluorescence was collected from 598 to 644 nm.
Accession Numbers
The accession numbers of the amino acid sequences of SRKb and other SRKs used in sequence alignments can be found in Supplemental Table 1 and Supplemental Data Set 2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Number of Potential N-Glycosylation Sites in the Extracellular Domains of SRKs from A. lyrata, A. halleri, B. oleracea, and B. rapa.
Supplemental Table 2. Distribution and Conservation of Potential N-Glycosylation Sites in the Extracellular Domains of SRKs from A. lyrata, A. halleri, B. oleracea, and B. rapa.
Supplemental Table 3. Primers Used for Plasmid Construction.
Supplemental Data Set 1. Amino Acid Sequence Alignment of SRK Variants from Arabidopsis lyrata (Al-SRKx), A. halleri (Ah-SRKx), Brassica oleracea (Bo-SRKx), and B. rapa (Br-SRKx).
Supplemental Data Set 2. Amino Acid Sequence Alignment of the PAN_APPLE Domains of SRK Variants from A. lyrata (Al-SRKx) and A. halleri (Ah-SRKx) SRK Variants.
Supplementary Material
Acknowledgments
We thank Tiffany Crispell for technical assistance and Anne Rea for generating the AtS1pro:mC-ER transgenic line. Description of this line and a detailed analysis of AtS1pro:SRKb-cYFP transformants for imaging of the plasma membrane-localized SRKb-cYFP protein were reported by Rea (2012). We thank Natasha Raikhel for the cYFP-containing plasmid, Hong-Gu Kang for the anti-H+-ATPase antibodies, and Shuh-ichi Nishikawa for the anti-BiP antibodies. The results described in this study are based upon work supported by the National Science Foundation (http://www.nsf.gov) under Grant IOS-1146725 to J.B.N. and M.E.N.
AUTHOR CONTRIBUTIONS
M.Y., T.T., M.E.N., and J.B.N. designed and performed the experiments and analyzed the data. M.Y., T.N., and J.B.N. wrote and revised the article.
Glossary
- SI
self-incompatibility
- PM
plasma membrane
- ER
endoplasmic reticulum
- Endo H
Endoglycosidase H
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Benham A.M. (2012). Protein secretion and the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 4: a012872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs N.A., Dwyer K.G., Nasrallah M.E., Nasrallah J.B. (2009a). In vivo detection of residues required for ligand-selective activation of the S-locus receptor in Arabidopsis. Curr. Biol. 19: 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs N.A., Nasrallah J.B., Nasrallah M.E. (2009b). Independent S-locus mutations caused self-fertility in Arabidopsis thaliana. PLoS Genet. 5: e1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P.Y., Fasman G.D. (1978). Empirical predictions of protein conformation. Annu. Rev. Biochem. 47: 251–276. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Dwyer K.G., Kandasamy M.K., Mahosky D.I., Acciai J., Kudish B.I., Miller J.E., Nasrallah M.E., Nasrallah J.B. (1994). A superfamily of S locus-related sequences in Arabidopsis: diverse structures and expression patterns. Plant Cell 6: 1829–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts I., Villmann C., Hollmann M. (1997). N-Glycosylation is not a prerequisite for glutamate receptor function but is essential for lectin modulation. Mol. Pharmacol. 52: 861–873. [DOI] [PubMed] [Google Scholar]
- Gavel Y., von Heijne G. (1990). Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng. 3: 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häweker H., Rips S., Koiwa H., Salomon S., Saijo Y., Chinchilla D., Robatzek S., von Schaewen A. (2010). Pattern recognition receptors require N-glycosylation to mediate plant immunity. J. Biol. Chem. 285: 4629–4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollecker M. (1997). Counting integral numbers of residues by chemical modification. In Protein Structure: A Practical Approach, Creighton T.E., ed (New York: IRL Press; ), pp. 151–164. [Google Scholar]
- Hong Z., Jin H., Tzfira T., Li J. (2008). Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20: 3418–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Wada I., Natsuka Y., Nagata K. (2006). EDEM accelerates ERAD by preventing aberrant dimer formation of misfolded alpha1-antitrypsin. Genes Cells 11: 465–476. [DOI] [PubMed] [Google Scholar]
- Jin H., Hong Z., Su W., Li J. (2009). A plant-specific calreticulin is a key retention factor for a defective brassinosteroid receptor in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 106: 13612–13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Yan Z., Nam K.H., Li J. (2007). Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol. Cell 26: 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo A., Schopfer C.R., Nasrallah M.E., Nasrallah J.B. (2001). Allele-specific receptor-ligand interactions in Brassica self-incompatibility. Science 293: 1824–1826. [DOI] [PubMed] [Google Scholar]
- Kandasamy M.K., Paolillo D.J., Faraday C.D., Nasrallah J.B., Nasrallah M.E. (1989). The S-locus specific glycoproteins of Brassica accumulate in the cell wall of developing stigma papillae. Dev. Biol. 134: 462–472. [DOI] [PubMed] [Google Scholar]
- Kho Y.O., Baër J. (1968). Observing pollen tubes by means of fluorescence. Euphytica 17: 298–302. [Google Scholar]
- Koncz C., Schell J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204: 383–396. [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- Larsson, C.H. (1985). Plasma membranes. In Modern Methods in Plant Analysis, New Series, Vol. 1, Cell Components, H.F. Linskens and J.F. Jackson, eds (Berlin: Springer-Verlag), pp. 85–104. [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lee H.Y., Bowen C.H., Popescu G.V., Kang H.G., Kato N., Ma S., Dinesh-Kumar S., Snyder M., Popescu S.C. (2011). Arabidopsis RTNLB1 and RTNLB2 Reticulon-like proteins regulate intracellular trafficking and activity of the FLS2 immune receptor. Plant Cell 23: 3374–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre B., Klaus-Heisen D., Pietraszewska-Bogiel A., Hervé C., Camut S., Auriac M.C., Gasciolli V., Nurisso A., Gadella T.W., Cullimore J. (2012). Role of N-glycosylation sites and CXC motifs in trafficking of Medicago truncatula Nod factor perception protein to plasma membrane. J. Biol. Chem. 297: 10812–10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Sherman-Broyles S., Nasrallah M.E., Nasrallah J.B. (2007). A cryptic modifier causing transient self-incompatibility in Arabidopsis thaliana. Curr. Biol. 17: 734–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maley F., Trimble R.B., Tarentino A.L., Plummer T.H. Jr (1989). Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal. Biochem. 180: 195–204. [DOI] [PubMed] [Google Scholar]
- Mellquist J.L., Kasturi L., Spitalnik S.L., Shakin-Eshleman S.H. (1998). The amino acid following an asn-X-Ser/Thr sequon is an important determinant of N-linked core glycosylation efficiency. Biochemistry 37: 6833–6837. [DOI] [PubMed] [Google Scholar]
- Mezghrani A., Fassio A., Benham A., Simmen T., Braakman I., Sitia R. (2001). Manipulation of oxidative protein folding and PDI redox state in mammalian cells. EMBO J. 20: 6288–6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naithani S., Chookajorn T., Ripoll D.R., Nasrallah J.B. (2007). Structural modules for receptor dimerization in the S-locus receptor kinase extracellular domain. Proc. Natl. Acad. Sci. USA 104: 12211–12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah M.E., Liu P., Sherman-Broyles S., Boggs N.A., Nasrallah J.B. (2004). Natural variation in expression of self-incompatibility in Arabidopsis thaliana: implications for the evolution of selfing. Proc. Natl. Acad. Sci. USA 101: 16070–16074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V., et al. (2009). Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 28: 3428–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Nishio T., Kusaba M. (2000). Sequence diversity of SLG and SRK in Brassica oleracea. Ann. Bot. (Lond.) 85: 141–146. [Google Scholar]
- Ohtsubo K., Marth J.D. (2006). Glycosylation in cellular mechanisms of health and disease. Cell 126: 855–867. [DOI] [PubMed] [Google Scholar]
- Rea A.C. (2012). In Vivo Imaging of the S-Locus Receptor Kinase in Transgenic Self-Incompatible Arabidopsis thaliana. PhD dissertation (Ithaca, NY: Cornell University). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruddock L.W., Molinari M. (2006). N-glycan processing in ER quality control. J. Cell Sci. 119: 4373–4380. [DOI] [PubMed] [Google Scholar]
- Saijo Y. (2010). ER quality control of immune receptors and regulators in plants. Cell. Microbiol. 12: 716–724. [DOI] [PubMed] [Google Scholar]
- Sainudiin R., Wong W.S.W., Yogeeswaran K., Nasrallah J.B., Yang Z., Nielsen R. (2005). Detecting site-specific physicochemical selective pressures: applications to the Class I HLA of the human major histocompatibility complex and the SRK of the plant sporophytic self-incompatibility system. J. Mol. Evol. 60: 315–326. [DOI] [PubMed] [Google Scholar]
- Sakoh-Nakatogawa M., Nishikawa S., Endo T. (2009). Roles of protein-disulfide isomerase-mediated disulfide bond formation of yeast Mnl1p in endoplasmic reticulum-associated degradation. J. Biol. Chem. 284: 11815–11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker R.H., Elleman C.J., Dickinson H.G. (1988). Control of pollen hydration in Brassica requires continued protein synthesis, and glycosylation in necessary for intraspecific incompatibility. Proc. Natl. Acad. Sci. USA 85: 4340–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slieker L.J., Martensen T.M., Lane M.D. (1986). Synthesis of epidermal growth factor receptor in human A431 cells. Glycosylation-dependent acquisition of ligand binding activity occurs post-translationally in the endoplasmic reticulum. J. Biol. Chem. 261: 15233–15241. [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderquist A.M., Carpenter G. (1984). Glycosylation of the epidermal growth factor receptor in A-431 cells. The contribution of carbohydrate to receptor function. J. Biol. Chem. 259: 12586–12594. [PubMed] [Google Scholar]
- Stein J.C., Dixit R., Nasrallah M.E., Nasrallah J.B. (1996). SRK, the stigma-specific S locus receptor kinase of Brassica, is targeted to the plasma membrane in transgenic tobacco. Plant Cell 8: 429–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Cao Y., Jansen Labby K., Bittel P., Boller T., Bent A.F. (2012). Probing the Arabidopsis flagellin receptor: FLS2-FLS2 association and the contributions of specific domains to signaling function. Plant Cell 24: 1096–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S., Isogai A., Tsukamoto C., Shiozawa H., Ueda Y., Hinata K., Okazaki K., Koseki K. (1989). Structures of N-glycosidic saccharide chains in S-glycoproteins, products of S-genes associated with self-incompatibility in Brassica campestris. Agric. Biol. Chem. 53: 713–722. [Google Scholar]
- Takayama S., Shimosato H., Shiba H., Funato M., Che F.S., Watanabe M., Iwano M., Isogai A. (2001). Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature 413: 534–538. [DOI] [PubMed] [Google Scholar]
- Tokhtaeva E., Munson K., Sachs G., Vagin O. (2010). N-glycan-dependent quality control of the Na,K-ATPase beta(2) subunit. Biochemistry 49: 3116–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76: 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach A.L., Lalonde B.A., Kandasamy M.K., Nasrallah J.B., Nasrallah M.E. (1990). Immunodetection of protein glycoforms encoded by two independent genes of the self-incompatibility multigene family of Brassica. Plant Physiol. 93: 739–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn R.A.L., Wulff B.B.H., Rivas S., Durrant M.C., van der Ploeg A., de Wit P.J.G.M., Jones J.D.G. (2005). Structure-function analysis of cf-9, a receptor-like protein with extracytoplasmic leucine-rich repeats. Plant Cell 17: 1000–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Maruyama D., Endo T., Nishikawa S. (2008). Arabidopsis thaliana has a set of J proteins in the endoplasmic reticulum that are conserved from yeast to animals and plants. Plant Cell Physiol. 49: 1547–1562. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.