Figure 5.
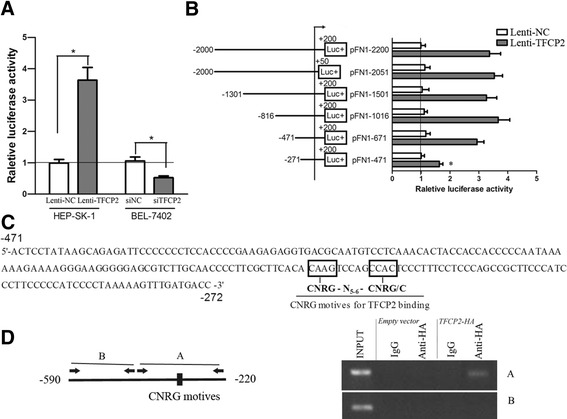
Deletional analysis of the FN1 minimal promoter, and analysis of TFCP2-binding sites. (A) FN1 promoter activity in SK-HEP-1 and BEL-7402 cells after the alteration of TFCP2. Firefly luciferase activity representing FN1 promoter activity was measured after transfection with pFN1-2200 and normalized to Renilla luciferase activity. (B) Schematic representation of the FN1 promoter (and its fragmental constructs) and transcription activation of FN1 promoter fragments by TFCP2. SK-HEP-1 control and overexpression cells were transfected with the deletion fragmental constructs of the FN1 promoter and renilla TK plasmid for normalization. All determinations were performed at least in triplicate in three independent experiments. Values represent mean ± SD. *P < 0.01. (C) The −471/-272 sequence of the FN1 promoter includes CNRG-N5–6-CNRG/C, as a putative site for TFCP2 binding. The positions of potential regulatory motifs are indicated by rectangles. Consensus sequence is annotated, where N = T, C, A or G; and R = A or G. (D) Schematic diagram of FN1 promoter showing the location of potential TFCP2-binding sites in the promoter and primers designed for ChIP assay. Fragment A and B represent fragments with potential TFCP2-binding consensus or without, respectively. ChIP using anti-IgG or anti-HA were performed with stable SK-HEP-1 cell lines expressing TFCP2-HA or not (empty vector) followed by PCR using primers to the candidate target promoter regions of FN1.
