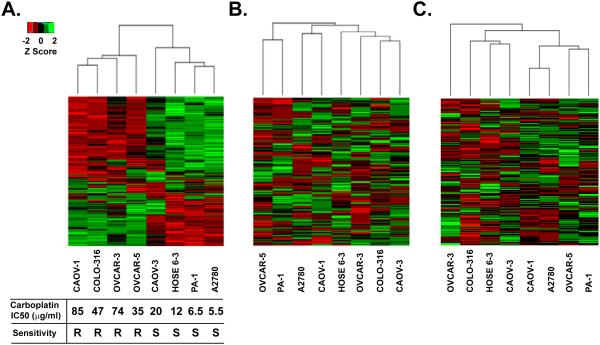
Figure 3. Hierarchical clustering of iTraq z-scores for ovarian cancer cell lines.
(A) The top 300 most variable iTRAQ proteins determined by median absolute deviation were clustered using the Eculidean complete method for the indicated ovarian cancer cell lines. Minimum z-scores are depicted in red and maximum z-scores are in green. Each row represents an individual protein, each column is the sum of the proteins identified in the indicated cell line. Carboplatin survival curves were determined by performing MTT cell viability assay and plotted for the 7 epithelial ovarian cancer cell lines and one control cell line. Absorbance was converted to the percentage of cells surviving and plotted against the concentration to calculate the IC50. Carboplatin-resistant cell lines were labeled “R” and cell lines sensitive to carboplatin treatment were labeled “S”. (B) Unsupervised hierarchical clustering of the same 300 most variable iTRAQ proteins using mRNA expression data. (C) Unsupervised hierarchical clustering of the same 300 most variable iTRAQ proteins using DNA methylation data.