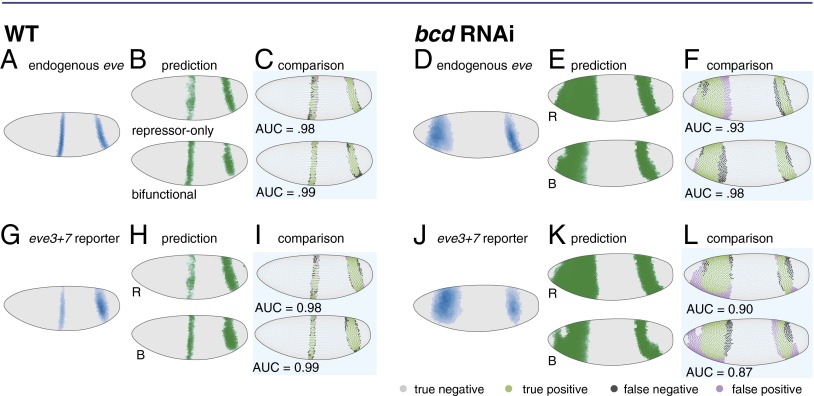
Fig. 3.
In bcd RNAi embryos the bifunctional model more accurately predicts the endogenous pattern and the repressor-only model more accurately predicts the eve3+7 reporter pattern. (A) The endogenous eve pattern in WT embryos is shown as a rendering of a gene expression atlas. Cells with expression below an ON/OFF threshold (Materials and Methods) are plotted in gray. For cells above this threshold, darker color indicates higher relative amounts. (B) The predictions of the repressor-only (R) and bifunctional (B) models in WT embryos. (C) Comparison of model predictions to the endogenous pattern in WT embryos. Green cells are true positives, purple cells are false positives, dark gray cells are false negatives, and light gray cells are true negatives. For visualization the threshold is set to 80% sensitivity, but the AUC metric quantifies performance over all thresholds. (D) The endogenous eve pattern in bcd RNAi embryos. (E) The predictions of the repressor-only (R) and bifunctional (B) models in bcd RNAi embryos. (F) Comparison of model predictions to the endogenous pattern in bcd RNAi embryos. The bifunctional model more accurately predicts the endogenous pattern in bcd RNAi embryos. (G–L) Same as A–F, respectively, for the eve3+7 reporter pattern. The repressor-only model predicts the eve3+7 reporter pattern more accurately in bcd RNAi embryos. Model parameters and AUC scores are in Tables S1 and S2.