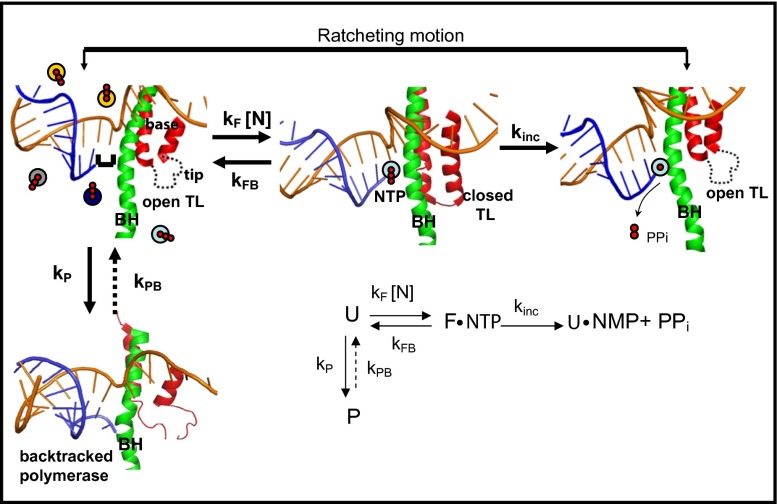
Fig. 5.
Diagram of structural changes that occur during elongation. Template DNA is shown in gold, RNA in blue; the TL structure is shown in red and the BH in green. (From Left to Right) The complex is in a posttranslocated position in which the NTP binding site is available for the next NTP to bind, and the polymerase is in an open conformation with an unfolded TL. Here the colored circles represent the four types of NTPs in solution. Next, once NTP binds, the TL folds into its double helical structure putting the polymerase in a closed conformation, which blocks the secondary channel and correctly positions the incoming NTP so incorporation can occur. After NTP incorporation is complete, the resulting PPi is released, the TL unfolds and the polymerase again adopts an open conformation. After nucleotide incorporation, the polymerase is in a pretranslocated state, and movements of the BH and the unfolded TL then take the polymerase to the posttranslocated state for the start of another cycle. The diagram also shows the pausing pathway, in which the TL is partially unfolded and the 3′ end of the RNA is displaced from the catalytic site of the enzyme. The structures used are based on the T. Thermopilus crystal structures in the absence [Protein Data Bank (PDB) ID code 2O5I] and presence (PDB ID code 2O5J) of NTP, and the paused state is based on the Pol II backtracked structure (PDB ID code 3GTJ). (Inset) Simplified kinetic scheme.