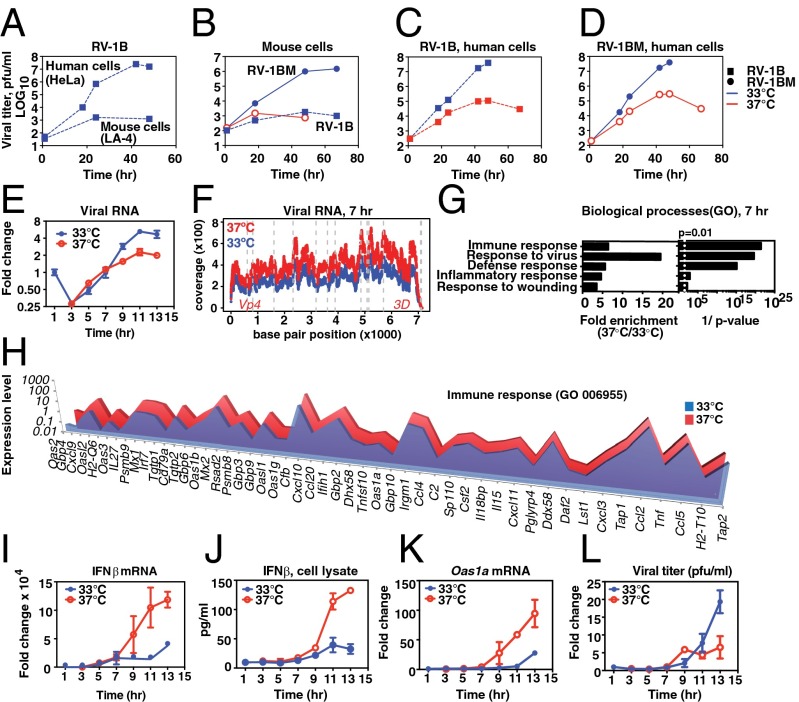
Fig. 1.
Temperature-dependent replication of rhinovirus and host response. (A–D) Cells were inoculated with a multiplicity of infection (MOI) of 0.001 of the indicated virus, and then incubated at 33 °C (blue line) or at 37 °C (red line). (A) At 33 °C, RV-1B exhibited ∼105-fold amplification in the human cell line, HeLa (upper line) but <50-fold increase in titer in mouse LA-4 cells (lower line). (B) Mouse-adapted RV-1B, RV-1BM, displayed ∼104-fold amplification in LA-4 cells at 33 °C (circles, solid line) compared to the minimal replication of RV-1B (squares, dashed line). At 37 °C, RV-1BM replicated less than 50-fold (solid red line) and RV-1B replication was not observed (not shown). (C and D) Growth curve of RV-1BM and RV-1B in HeLa cells at 33 °C (blue line) or at 37 °C (red line). (E–L) Primary mouse AECs were inoculated with RV-1BM, MOI 20, and incubated at 33 °C or 37 °C following the initial 1-h inoculation at 33 °C. (E) Fold change in viral RNA. (F) RNA-Seq results showing representation of the RNA viral genome at 7 h postinfection in cells incubated at 37 °C vs. 33 °C. The y axis shows the coverage at each position: (number of reads at each position/total number of mapped reads in the sample) × 106; the x axis represents the position in the viral genome. (G and H) DAVID analysis of host mRNAs differentially enriched during RV-1BM infection at 37 °C compared with 33 °C. (G) Transcripts that differed in expression by at least twofold were included in the analysis (364 transcripts). Gene Ontology (GO) database clusters up-regulated at 37 °C (P < 0.01) are shown with fold enrichment (Left) and Bonferroni-corrected P values (Right). (H) For the immune response cluster, genes differentially expressed by greater than twofold are shown, arranged from greatest (left) to least (right) differential expression. (I) Fold change in host cell IFN-β mRNA during the single-step RV-1BM infection shown in E, normalized to Hprt. (J) IFN-β protein detected by ELISA in cell lysates prepared from replicate cultures of the single-step growth curve experiment shown in E. (K) Fold change in Oas1a mRNA during the single-step RV-1BM infection shown in E. (L) Viral titers of cell lysates prepared from replicate cultures of the single-step growth curve experiment shown in E were determined by plaque assay. Points and error bars represent the mean and SEM of two or three replicates per condition. Data are representative of at least three independent experiments.