Significance
DNA methylation, a chemical mark on chromatin, while not affecting DNA's primary sequence, plays important roles in silencing “bad DNA” that would become deleterious to cells if abnormally expressed. This DNA methylation-mediated silencing system against bad DNA is tightly regulated to prevent the misplacement of methylation on “good DNA.” In Arabidopsis thaliana, DOMAINS REARRANGED METHYLTRANSFERASE2 (DRM2) controls RNA-directed DNA methylation in a pathway that also involves the plant-specific RNA Polymerase V (Pol V). The Arabidopsis genome also encodes an evolutionarily conserved but catalytically inactive methyltransferase, DRM3. Here, we investigate the molecular mechanism of DRM3 action on DNA methylation and its dynamic regulation of Pol V transcription. Together, this study sheds further light on the mechanism of RNA-directed DNA methylation.
Keywords: DNA methylation, epigenetic regulation, RNA polymerase, non-coding RNA, gene silencing
Abstract
DNA methylation is a mechanism of epigenetic gene regulation and genome defense conserved in many eukaryotic organisms. In Arabidopsis, the DNA methyltransferase DOMAINS REARRANGED METHYLASE 2 (DRM2) controls RNA-directed DNA methylation in a pathway that also involves the plant-specific RNA Polymerase V (Pol V). Additionally, the Arabidopsis genome encodes an evolutionarily conserved but catalytically inactive DNA methyltransferase, DRM3. Here, we show that DRM3 has moderate effects on global DNA methylation and small RNA abundance and that DRM3 physically interacts with Pol V. In Arabidopsis drm3 mutants, we observe a lower level of Pol V-dependent noncoding RNA transcripts even though Pol V chromatin occupancy is increased at many sites in the genome. These findings suggest that DRM3 acts to promote Pol V transcriptional elongation or assist in the stabilization of Pol V transcripts. This work sheds further light on the mechanism by which long noncoding RNAs facilitate RNA-directed DNA methylation.
In eukaryotes, DNA methylation plays significant roles in gene silencing and controls many important biological processes, including genome imprinting (1), X chromosome inactivation (2), genome stability, and the silencing of transposons, retroviruses, and other harmful DNA elements (3–5). In Arabidopsis, DNA methylation occurs in CG, CHG, and CHH (where H = A, T, or C) sequence contexts and is controlled by at least four DNA methyltransferases: METHYLTRANSFERASE 1 (MET1), CHROMOMETHYLASE2 (CMT2), CHROMOMETHYLASE3 (CMT3), and DOMAINS REARRANGED METHYLASE 2 (DRM2). MET1 and DRM2 are the plant homologs of mammalian methyltransferases Dnmt1 and Dnmt3, respectively, whereas CMT2 and CMT3 are plant-specific methyltransferases. MET1, like Dnmt1, is important for maintenance of CG methylation during DNA replication (6–8), and DRM2, CMT3, and CMT2 control the maintenance of non-CG methylation (9–13). The establishment of DNA methylation is controlled by DRM2 (9) via a process termed RNA-directed DNA methylation (RdDM) (14–16).
In the current RdDM model, the DNA-DEPENDENT RNA POLYMERASE IV (Pol IV), RNA-DEPENDENT RNA POLYMERASE 2, and DICER-LIKE 3 function together to produce small interfering RNAs (siRNAs) that are bound by ARGONAUTE4 (AGO4). The AGO4/siRNA complex is thought to base pair with noncoding RNA transcripts produced by a DNA-DEPENDENT RNA POLYMERASE V (Pol V). Production of Pol V transcripts, as well as genome-wide Pol V chromatin occupancy, requires a complex termed DDR consisting of the putative chromatin-remodeling factor DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1 (DRD1), structural maintenance of chromosome domain protein DEFECTIVE IN MERISTEM SILENCING 3 (DMS3), and RNA-DIRECTED DNA METHYLATION 1 (17–20). The SUPPRESSOR OF VARIEGATION 3–9 HOMOLOG 2 (SUVH2) and -9 (SUVH9) proteins are required for genome-wide Pol V chromatin association by binding to DNA methylation (21, 22). The cooccurrence of Pol IV-dependent siRNAs, Pol V-dependent noncoding RNA transcripts, and AGO4–DRM2 interaction (23) is thought to ultimately guide DRM2 to specific genomic sequences to cause DNA methylation.
The Arabidopsis genome encodes another DNA methyltransferase-like gene, DOMAINS REARRANGED METHYLTRANSFERASE 3 (DRM3), that is homologous to DRM2. Similar to DRM2, DRM3 has N-terminal ubiquitin-associated (UBA) domains and a C-terminal methyltransferase domain (24). Sequence alignments of the DRM3 methyltransferase domains from many plant species revealed the absence of highly conserved key amino acids known to be critical for catalytic activity. However, DRM3 is required for full levels of DRM2-mediated DNA methylation (24, 25). Similarly, mammalian genomes encode a catalytically inactive methyltransferase termed Dnmt3L that also lacks catalytically critical residues but is required for de novo DNA methylation in vivo (26–29). Dnmt3L forms a hetero-tetrameric complex with Dnmt3a, and the interaction of Dnmt3a with Dnmt3L stimulates its activity (27–29). Despite this similarity, DRM3 and Dnmt3L differ significantly in their N-terminal domains. Dnmt3L has an N-terminal ADD domain that specifically recognizes histone 3 unmethylated at lysine 4 and targets Dnmt3 to chromatin (30, 31). In contrast, DRM3 has N-terminal UBA domains of unknown function (23, 24). Despite the importance of DRM3 for DRM2-mediated DNA methylation, it is unclear whether DRM3 acts similarly to mammalian Dnmt3L by interacting with DRM2 and stimulating DRM2 activity. It is also unknown whether DRM3 is a general or a locus-specific factor required for DNA methylation. Furthermore, the relationship between DRM3 and other components in the RdDM pathway remains elusive.
Here, through a combination of genetic, genomic, and biochemical approaches, we describe aspects of the molecular mechanism of DRM3 action in Arabidopsis. We show that DRM3 acts as a general factor controlling global DNA methylation and small RNA abundances and interacts physically with Pol V. To further understand the molecular basis of the DRM3-Pol V interaction, we determined Pol V genome-wide chromatin occupancy in drm3 and found surprisingly that DRM3 target sites have either gain or loss of Pol V in drm3. Similar to drm2, the sites that lost Pol V occupancy in drm3 had relatively low methylation levels, siRNAs, and cytosine contents, suggesting that DNA methylation is important for retaining Pol V at these sites. This methylation-dependent Pol V retention is consistent with our recent observation in met1 (CG methyltransferase) that loss of DNA methylation results in loss of Pol V chromatin association (21). We also observed that many additional sites gain Pol V occupancy in drm3. However, despite gaining Pol V occupancy, we observed a reduction in the abundance of Pol V-dependent transcripts in drm3, suggesting that DRM3 may stabilize Pol V transcripts and/or mediate Pol V elongation. Together, our results indicate that DRM3 controls DNA methylation through its functional interaction with Pol V and by regulating Pol V transcript levels.
Results and Discussion
DRM3 Is a General Factor That Has Moderate Effects on DNA Methylation at RdDM Targets.
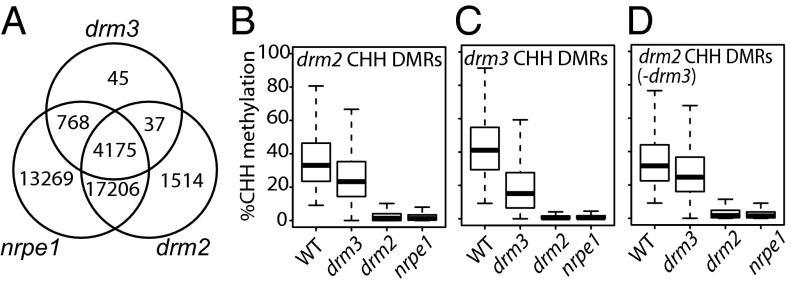
A previous large-scale DNA methylome study of 86 Arabidopsis gene-silencing mutants revealed that genome-wide DNA methylation is partially reduced in drm3 mutant (25). To test whether DRM3 acts as a general or specific factor for genome-wide DNA methylation, we performed an extensive analysis of the bisulfite-sequencing (BS-seq) data in drm3, as well as that of drm2 and nrpe1 mutants (the catalytic subunit of Pol V) for comparison. We first defined differentially methylated regions (DMRs) in each of these mutants for the CHH context because the RdDM pathway primarily impacts CHH methylation (25). We observed that the majority of drm3 DMRs (∼98%) overlap with DMRs of nrpe1 (Fig. 1A), suggesting that DRM3 acts mainly at RdDM target sites. Consistently, we noted a strong overlap in the DMRs defined in drm2 and nrpe1 mutants although there were a greater number of DMRs in nrpe1 than in drm2, which is likely due to the higher sequencing depth of the nrpe1 library relative to drm2 (Materials and Methods). Additionally we observed many fewer DMRs in drm3 than in drm2 (Fig. 1A). It is possible that DRM3 specifically acts at certain RdDM targets. Alternatively, DRM3 might have a generally weak methylation defect at most RdDM targets, in which case drm3-specific DMRs might be mainly due to significance cutoff effects we impose in the DMR calling procedure. Toward this end, we determined the percent methylation in each cytosine context (CG, CHG, CHH) across drm2 DMRs and noted a moderate loss of DNA methylation in drm3 (Fig. 1B and Fig. S1A). We also quantified DNA methylation patterns at drm3 CHH DMRs and showed that drm3 mutants had a stronger effect at drm3 CHH DMRs than those of all drm2 DMRs (Fig. 1C). However, even at drm3 CHH DMRs, the effects of the drm2 and nrpe1 mutations were much stronger than those of the drm3 mutation (Fig. 1C). Furthermore, at drm2 CHH DMRs that were not called as drm3 DMRs, we still noted a small but significant effect (P < 2.2e−16; Wilcoxon signed rank test) of the drm3 mutation on DNA methylation (Fig. 1D). Together, these analyses suggest that DRM3 is a weak RdDM factor acting at most sites, rather than a locus-specific methylation factor.
Fig. 1.
DRM3 has moderate effects on global DNA methylation at RNA-directed DNA methylation targets. (A) Overlap of differentially methylated regions (DMRs) identified in drm3, drm2, and nrpe1 mutants. CHH methylation levels over (B) drm2 hypo-DMRs, (C) drm3 hypo-DMRs, and (D) drm2 hypo-DMRs excluding drm3 hypo-DMRs in WT, drm2, nrpe1, and drm3. The CHH methylation in drm3 is significantly reduced compared with WT (P < 2.2e−16; Wilcoxon signed rank test). CHH methylation levels in drm2 and nrpe1 are both significantly reduced compared with drm3 (P < 2.2e−16; Wilcoxon signed rank test).
DRM3 Acts Downstream of the Production of siRNAs.
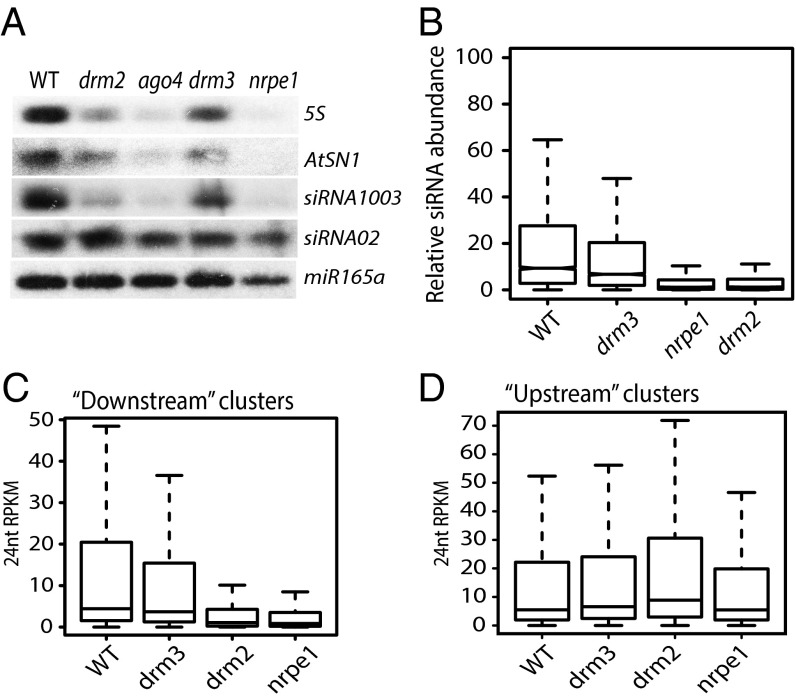
RdDM has two main phases, siRNA production and methylation targeting. To place DRM3 in the RdDM pathway, we analyzed the accumulation of siRNAs in the drm3 mutants. We first assessed the abundance of siRNAs derived from the 5S rDNA, AtSN1, siRNA 02, and siRNA 1003 loci in drm3 by Northern blotting. The 5S rDNA, AtSN1, and siRNA 1003 were shown previously to be dependent on Pol IV and Pol V (32) whereas siRNA 02 was shown to be dependent only on Pol IV (32), but unaffected in downstream effector mutants such as nrpe1, ago4, and drm2. Consistent with a previous report (24), we observed partially reduced accumulation of siRNAs at 5S rDNA, AtSN1, and siRNA 1003, but not at siRNA 02 (Fig. 2A), suggesting that DRM3 likely acts in the downstream portion of the RdDM pathway. To further place DRM3 within the RdDM pathway, we generated siRNA libraries and performed high-throughput sequencing from WT, nrpe1, drm2, and drm3. Consistent with the partial reduction of DNA methylation levels, we found that the abundance of siRNAs in drm3 was also slightly but significantly reduced relative to WT at drm2 DMRs (Fig. 2B and Fig. S1B). Furthermore, using small RNA clusters previously defined as being affected only in the upstream mutants versus clusters affected in downstream mutants (33), we were able to see a significant loss of 24 nucleotide small RNAs at the downstream clusters (Fig. 2C) (P = 4.611e−10; Wilcoxon signed rank test) but not at the upstream clusters in drm3 mutants (Fig. 2D). Together, these data suggest that DRM3 acts in the downstream part of the RdDM pathway.
Fig. 2.
DRM3 acts downstream of the production of siRNAs. (A) Northern blots show accumulation levels of small RNAs at 5S rDNA, AtSN1, and siRNA1003 loci. The siRNA02 and miR165a serve as two internal controls. (B) Average distribution of 24-nt siRNA reads over drm2 CHH hypo-DMRs. The siRNA abundance is significantly reduced compared with WT in all mutants (P < 2.2e−16; Wilcoxon signed rank test). Levels of 24-nt siRNAs in WT and mutants over previously defined siRNA clusters affected in the “downstream” (C) and “upstream” (D) RdDM mutants. The 24-nt siRNA levels are only significantly reduced in drm3 (P = 4.611e−10; Wilcoxon signed rank test), drm2 (P < 2.2e−16; Wilcoxon signed rank test), and nrpe1 (P < 2.2e−16; Wilcoxon signed rank test) compared with those of WT in the downstream clusters (C).
DRM3 Interacts with Pol V.
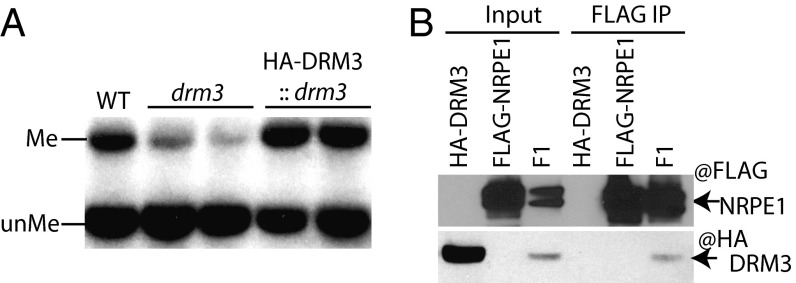
To gain further insights into the mechanism of DRM3 action, we purified an HA-epitope–tagged DRM3 expressed from a transgene introduced into Arabidopsis drm3 and under the control of DRM3 endogenous promoter (Fig. S2A). The complementation of the drm3 mutant by transgenic DRM3 was confirmed by methylation-sensitive enzyme digestion and Southern blot analysis at the MEA-ISR locus (Fig. 3A). After affinity purification, DRM3-associated proteins were identified by multidimensional protein identification technology mass spectrometry. Table S1 shows a partial list of copurifying proteins. We did not detect any DRM2 peptides in the mass spectrometry data (Table S1), nor were we able to detect DRM2 in coimmunoprecipitation (co-IP) assays with DRM3 (Fig. S2C). This failure detection of DRM3-DRM2 interaction suggests that DRM3 acts differently than its mammalian homolog Dnmt3L and does not interact with DRM2. Instead, we identified seven Pol V subunits with significant sequence coverage, including previously reported Pol V-specific subunits NRPE1, NRPE3B, and NRPE7, as well as Pol IV/Pol V-shared subunits NRPE2, NRPE3A, NRPE11, and NRPE9A (Table S1). To further confirm these interactions, we performed immunoprecipitation and mass spectrometric analysis of the Pol V complex purified from an Arabidopsis transgenic line expressing the FLAG-tagged Pol V largest subunit (NRPE1-FLAG). This analysis indeed detected DRM3 peptides in addition to all known Pol V subunits (Table S2). Consistent with previous published purifications (19, 34–36), we also identified other Pol V interactors, including AGO4, DMS3, DRD1, and KTF1/SPT5L. To further validate the mass spectrometry data, we performed co-IP experiments from F1 plants expressing both HA-tagged DRM3 and FLAG-tagged NRPE1. As shown in Fig. 3B, when we pulled down NRPE1 with anti-FLAG beads, we could detect DRM3 with an anti-HA antibody. This interaction was further confirmed by a reciprocal co-IP (Fig. S2B). We were unable to detect an interaction between DRM3 and AGO4, one of the other Pol V interactors, suggesting that DRM3 may form distinct complexes with Pol V, or that the weak association of both AGO4 and DRM3 with Pol V does not allow for detection of a very weak but indirect association of AGO4 and DRM3. In summary, our data suggest that DRM3 physically associates with the Pol V complex.
Fig. 3.
DRM3 is associated with Pol V in vivo. (A) Complementation test of drm3 mutant with pDRM3::HA-DRM3 at the MEA-ISR locus by restriction digestion with methylation-sensitive MspI and Southern blot. Me, methylated DNA; unMe, unmethylated DNA. (B) Coimmunoprecipitation assays confirming DRM3–NRPE1 interaction. Input lanes confirm the expression of the epitope-tagged proteins in the parental lines and F1. F1 represents the first generation from a cross between the two parental lines.
DRM3 Mediates Pol V Chromatin Association at Specific Target Sites.
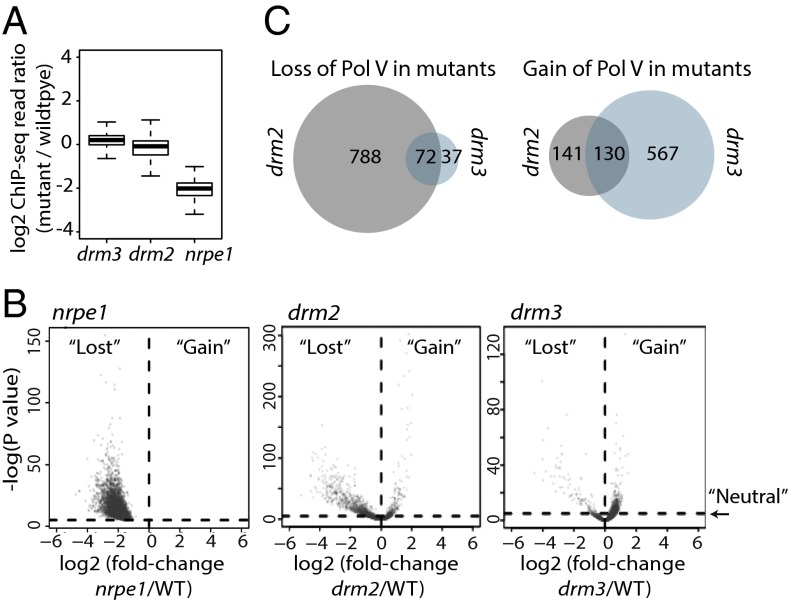
Given the association of DRM3 with the Pol V complex, we tested whether Pol V chromatin association might be affected in drm3 mutants in a manner similar to mutants in the DDR complex components (20). Thus, we performed chromatin immunoprecipitation followed by sequencing (ChIP-seq) of Pol V using an endogenous antibody against the NRPE1 subunit of Pol V in WT and drm3 plants. Additionally, ChIP-seq was performed on nrpe1 mutants as a negative control and drm2 mutants as a control for downstream RdDM mutants to enable detection of any specific effects of the drm3 mutation. Interestingly, using the endogenous Pol V antibody, we were able to identify >60% more high confidence Pol V binding sites (n = 4,317) from the resulting sequencing data than we previously observed using an epitope-tagged version of Pol V (20) (n = 2,656). The sequencing also revealed that neither drm2 nor drm3 mutants displayed large-scale loss of Pol V chromatin occupancy relative to WT, compared with the nrpe1 mutants (Fig. 4A). This observation is largely consistent with earlier studies showing that loss of DRM2 does not affect Pol V targeting (18). However, upon close inspection of the sequencing data, we found that certain subsets of Pol V targets experienced either loss or gain of Pol V in drm2 or drm3 mutants (Fig. 4B and Fig. S3 A and B). We also observed that the Pol V signal was increased at a larger number of sites in drm3 than those that were decreased (Fig. 4C). In drm2, the situation was reversed, with more sites losing than gaining Pol V (Fig. 4C). Interestingly, sites that gained Pol V in drm3 overlapped with sites that gained in drm2 and vice versa (Fig. 4C). We repeated ChIP-seq analysis of Pol V in drm3 mutants using the previously described FLAG-tagged NRPE1 transgene two more times and confirmed a similar alteration in patterns of Pol V chromatin occupancy that we observed with the endogenous Pol V antibody, and that drm3 mutants do not alter the overall levels of Pol V (Fig. S3 C and D). These data suggest that DRM2 and DRM3 dynamically regulate Pol V chromatin association and that drm3 mutants increase Pol V occupancy at many sites.
Fig. 4.
DRM2 and DMR3 differentially affect Pol V occupancy. (A) Distribution of log2 ratios of Pol V enrichment in mutant vs. WT over previously defined Pol V sites. The drm2 shows a significantly greater loss of Pol V compared with drm3 (P < 2.2e−16; Wilcoxon signed rank test), and nrpe1 shows a greater reduction than either drm2 or drm3 (P < 2.2e−16; Wilcoxon signed rank test). (B) Scatter plots of the relationship between Pol V ChIP-seq fold-change in nrpe1, drm2, and drm3 mutants relative to WT and the significance of that change (P value; Fisher’s exact test) at defined Pol V binding sites (n = 4,317). P value cutoff (P < 1e−5) used to define Pol V sites with either a neutral change or a significant gain or loss of Pol V for each mutant is represented by a horizontal dotted line. (C) Venn diagrams representing the overlaps of Pol V sites (total = 4,317) that lose or gain Pol V in drm2 and drm3 mutants.
Pol V Occupancy Is Correlated with Methylation Levels and siRNA Abundance.
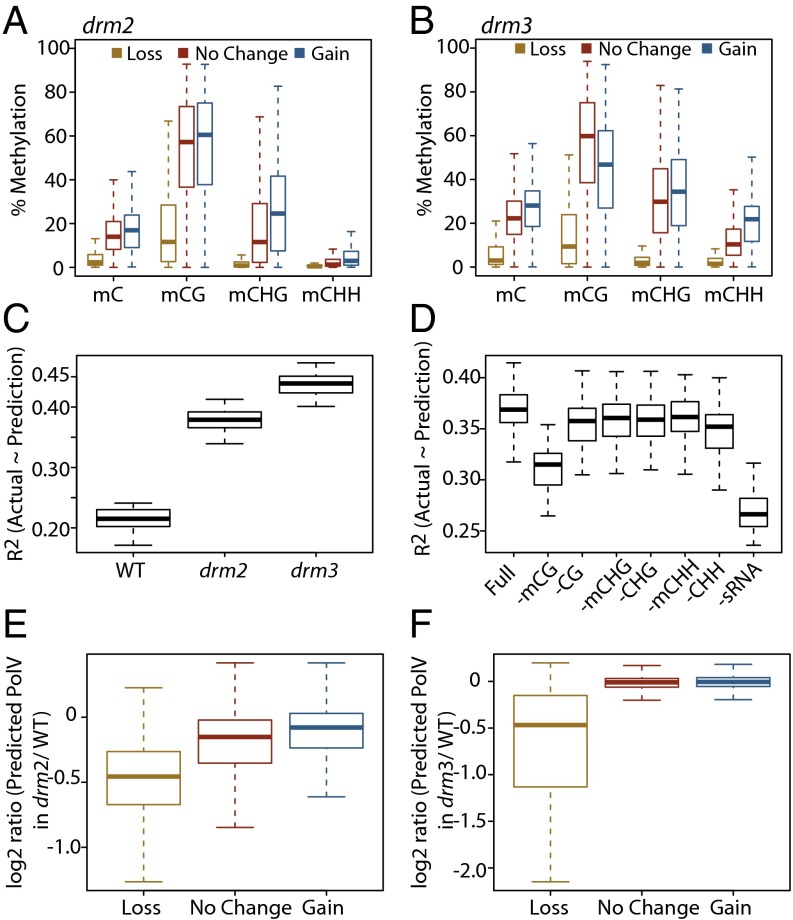
Pol V occupancy in the genome requires the methyl DNA binding proteins SUVH2 and SUVH9 and is almost completely lost in the strong DNA methylation mutant met1 (21). We therefore considered the possibility that alterations of DNA methylation patterns in drm2 or drm3 might mediate changes in Pol V chromatin association. To test this possibility, we analyzed the methylation levels in drm2 and drm3 mutants at Pol V sites that were classified as experiencing either loss, gain, or no change in Pol V occupancy. We found that sites that lost Pol V in drm2 had relatively low levels of methylation in all contexts in drm2 (yellow boxes in Fig. 5A). These sites were also the sites showing the greatest loss of DNA methylation in drm2 (yellow boxes in Fig. S4A). In contrast, Pol V sites that experienced little alteration (red boxes) or slight gains of Pol V occupancy (blue boxes) showed high levels of methylation in all sequence contexts (CG, CHG, and CHH) (Fig. 5A and Fig. S4A), suggesting that DNA methylation is a major factor driving retention of Pol V at chromatin consistent with our previous report (21). Similarly, our siRNA analyses revealed that the sites that did not lose Pol V occupancy in drm2 generally retained higher residual 24-nucleotide siRNA levels (Fig. S4B). Additionally, sites that lost Pol V occupancy in drm2 showed a lower cytosine content than sites that did not lose Pol V (Fig. S4C). Given this relationship between drm2 methylation, siRNAs, cytosine content, and Pol V occupancy, we hypothesized that the loss of Pol V chromatin association in drm3 mutants may be similarly due to an indirect effect of drm3-dependent alterations of DNA methylation and siRNAs. We therefore performed a similar analysis on drm3 mutants and found that altered levels of Pol V in drm3 mutants followed similar trends as the sites altered in drm2 mutants. Sites that lost Pol V in drm3 showed the lowest levels of methylation in all contexts in drm3 whereas sites that retained or gained Pol V tended to show higher levels of methylation (Fig. 5B). Moreover, sites showing loss of Pol V in drm3 showed lower starting levels of methylation and cytosines and showed a greater loss of methylation and siRNAs in drm3 than those sites that retained or gained Pol V (Fig. S4 D–F). Together, these results indicate that, in the absence of DRM2 or DRM3, Pol V targeting is largely dictated by sequence composition as well as the remaining DNA methylation and siRNAs.
Fig. 5.
Pol V association with chromatin in drm mutants is correlated to DNA methylation levels. (A) Methylation levels for different cytosine contexts in drm2 at Pol V sites classified as experiencing a “loss,” “gain,” or “no change” in Pol V occupancy in drm2. (B) Same representation as A, except methylation levels in a drm3 mutant are shown, and the Pol V sites are classified as experiencing a loss, gain, or no change in Pol V occupancy in drm3. The methylation levels of total methylated C (mC) and methylated CG (mCG) are significantly reduced (P < 2.2e−16; Wilcoxon rank sum test) at loss sites compared with gain or no change sites for both drm2 and drm3. (C) Distribution of R2 values of the fit of predicted Pol V signal to the actual observed Pol V ChIP-seq signal using models built upon methylation, cytosine content, and sRNA data derived from each respective genotype. Models were trained using 3/4 of the defined Pol V peaks and tested against the remaining 1/4 of sites. Training and testing were repeated 25 times for each genotype. The WT R2 value is significantly less than both the drm2 and drm3 values (P = 5.96e−08; Wilcoxon signed rank test). (D) Distribution of R2 values of predicted versus actual Pol V ChIP-seq signal for models trained on drm2 data as in C using the full set of parameters (“Full”) or subtracting one parameter (−mC, removing methylation data for a given context; −C, removing cytosine abundance data for a given context; −sRNA, removing 24-nt sRNA data). Both the −mCG and −sRNA models perform significantly worse than the next worse model (−CHH, P = 5.96e−08; Wilcoxon signed rank test). (E) Distribution of log2 ratios between predicted Pol V ChIP-seq signal in drm2 versus predicted signal in WT at Pol V sites classified as “Loss,” “Gain,” or “no change” in Pol V enrichment in drm2. The model was trained using drm3 data (without mCHH or sRNAs) at Pol V sites, excluding drm2 “Loss” or “Gain” sites. (F) Analogous to E, except that sites tested were classified based on Pol V behavior in drm3 and the model was generated using drm2 data, excluding the drm3 “Loss” or “Gain” sites. The loss category is significantly lower than the no change category (P < 2.2e−16; Wilcoxon rank sum test) for both drm2 and drm3.
Modeling of Pol V Occupancy.
We attempted to develop predictive models of the relationship between DNA methylation, siRNAs, cytosine content, and Pol V occupancy in the drm2 and drm3 mutants. We found that models generated from the drm2 or drm3 methylome/siRNA profiles performed better in predicting Pol V occupancy levels than models generated using WT profiles (Fig. 5C and Fig. S5A), perhaps because DNA methylation or siRNAs become limiting for Pol V recruitment in these mutant backgrounds. By individually removing predictor variables, we noted that, of the three sequence contexts, only CG-context methylation strongly affected the predictive power of Pol V occupancy modeling (Fig. 5D). These results are consistent with the almost complete loss of Pol V occupancy previously observed in the met1 mutant (21) and provide additional evidence that CG methylation is critical for retaining Pol V at chromatin. Consistent with the strong role of maintenance methylation in retaining Pol V, we found that, even when we removed both CHH methylation and 24-nucleotide siRNA abundance data (two hallmarks of RdDM), we could predict Pol V occupancy levels reasonably well in drm2 and drm3 mutants (Fig. S5B).
Using Pol V occupancy models of drm3 data (CG and CHG methylation, and cytosine content), we could train new models that successfully predicted the loss of Pol V that was experimentally observed in drm2 mutants (Fig. 5E). Reciprocally, we could train models using drm2 data to successfully predict loss of Pol V in drm3 (Fig. 5F). However, we found that both models failed to predict the observed gain of Pol V in drm2 and drm3 mutants (Fig. 5 E and F). Together, these results reinforce our hypothesis that the loss of Pol V chromatin occupancy in drm2 and drm3 mutants can be explained, and predicted, by alterations of DNA methylation caused by drm2 and drm3 mutants.
DRM3 Is Partially Required for Pol V-Dependent Noncoding RNA Transcript Accumulation.
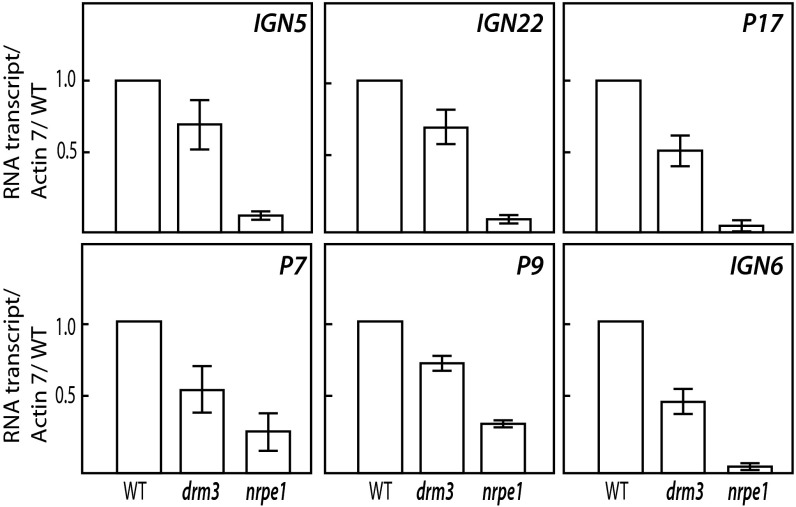
Although the loss of Pol V occupancy at certain sites in the drm3 mutant can be explained by an indirect effect of the loss of DNA methylation, we also observed that drm3 mutants show an increase of Pol V occupancy at a very large number of sites. The numbers of sites that gain Pol V in drm3 are much greater than the number of sites that gain Pol V in drm2 even though drm3 has a weaker effect on the RdDM than does drm2. Furthermore, the sites that gain Pol V in drm3 have lower levels of DNA methylation than those that gain Pol V in drm2 (Fig. S4), suggesting that the gains of Pol V in drm3 may not be simply driven by a redistribution of Pol V to highly methylated sites. The observations that drm3 shows losses of DNA methylation, and yet shows overall gains of Pol V occupancy, suggest that DRM3 acts at some point downstream of Pol V recruitment. Given the physical interaction of DRM3 with Pol V, we hypothesize that DRM3 might be needed for the efficient production of Pol V transcripts or for their stability. To test this hypothesis, we selected a number of represented Pol V-enriched sites that had either not lost or had gained Pol V occupancy in drm3 and assayed the Pol V RNA transcript levels in drm3 using quantitative reverse transcription PCR. We observed a partial but reproducible reduction of Pol V transcript at all tested sites (Fig. 6). One possible explanation for these results is that DRM3 may be involved in promoting efficient Pol V transcriptional elongation. This model might also explain the observed increase in Pol V occupancy because a more slowly transcribing Pol V would be expected to spend a longer time on chromatin, and thus give high signal strength in our chromatin immunoprecipitation experiments. A second possibility is that DRM3 might be needed for Pol V RNA transcript stability. Further in vitro Pol V transcription activity assays using a previously developed method (37) will be critical to test these possibilities. Together, these findings suggest a unique role of DRM3 in regulating the output of Pol V transcription activity and help to further elaborate the factors that are needed for proper functioning of the RdDM pathway.
Fig. 6.
DRM3 is partially required for Pol V-dependent noncoding RNA transcript accumulations. Quantitative RT-PCR analysis of the abundance of noncoding RNA transcripts from Pol V-enriched loci that either gained or did not lose Pol V occupancy in nrpe1 and drm3 mutants. Transcript levels were analyzed in WT, drm3, and nrpe1 plants and normalized to the levels of ACTIN7. Error bars represent the SD of more than five replicates.
Materials and Methods
Plant Material.
All Arabidopsis plants used in this study were in the Col-0 ecotype and were grown under long day conditions. The following Arabidopsis mutant lines were used: drm2-2 (SALK_150863), nrpe1-12 (SALK_033852), drm3-1 (SALK_136439), and ago4 (SALK_007523).
Bisulfite Sequencing Data Analysis.
The whole-genome BS-seq data from WT plants have been previously described (38), and whole genome datasets for drm3, drm2, and nrpe1 have also been described (25). The 50 mer sequencing reads were analyzed. Identical reads were collapsed into single reads, and reads were mapped to the TAIR10 genome using BSmap (39), allowing up to two mismatches and retaining only uniquely mapping reads. Fractional DNA methylation levels were computed by number of C (#C) #C/(#C + #T). DMRs were defined as previously described (25) with the following modifications: a false discovery rate (FDR) < 0.001 was required for a DMR to be called, and five cytosines with at least 5× coverage must be present in a candidate DMR. Called DMRs were not merged for downstream analysis.
RNA Analysis.
Total RNA was extracted from 0.5 g of flowers using TRIzol (Invitrogen). Small RNAs were purified as previously described (40). Small RNA Northern blot was performed as described previously (41). Small RNA libraries were generated and sequenced following the manufacturer’s instructions (Illumina). Adapter sequences were clipped off before mapping. Reads were mapped to the TAIR10 genome using Bowtie (42) by allowing no mismatches and keeping only reads that uniquely map to the genome. For the analyses, the small RNA counts were normalized to the size of each small RNA library by dividing the number of reads to the number of total uniquely mapping reads of 15–30 bp in size. For the comparison of “upstream” and “downstream” clusters the “pol-iv only” and “shh1/drm2/pol-v” clusters from ref. 33 were used, respectively.
Detection of Pol V-dependent transcripts was conducted as described in refs. 19 and 20. Total RNA was extracted from 0.1 g of flowers with TRIzol and treated with DNase I (Invitrogen). First-strand cDNA was synthesized with gene-specific primers (Table S3) using SuperScript III (Invitrogen). Quantification of transcripts was performed as described in ref. 19.
Generation of Epitope-Tagged DRM3 Transgenic Plants.
Epitope-tagged DRM3 constructs were generated as described previously (10). The full-length genomic DNA fragment containing a 3.5-kb promoter region was amplified by PCR (Table S3), and PCR products were cloned into the pCR2.1 vector (Invitrogen) with flanking Sal I sites. BamHI and ClaI sites were then introduced at the 5′ end of DRM3 by site-directed mutagenesis (200521; Stratagene). A 3xHA (HA = YPYDVPDYA) epitope tag was inserted using BamHI and ClaI sites. This tagged construct was then moved as a SalI fragment into pCAMBIA1300 binary vector and introduced into Agrobacterium strain AGLO, followed by transforming into drm3-1 mutant using the floral dip method (43). The third-generation transgenic plants were used for immunoprecipitation and mass spectrometry analyses.
Affinity Purification and Mass Spectrometry.
The immunoprecipitation (IP) was performed as previously described (10). The extracts from ∼10 g of flowers expressing HA-DRM3 or FLAG-NRPE1 were incubated with 200 µL of anti-HA affinity matrix (11815016001; Roche) and M2 Flag agarose beads (A2220; Sigma), respectively, at 4 °C for 2–3 h. The bead-bound complex was then washed two times with 40 mL of lysis buffer (LB) and four additional times with 1 mL of LB by mixing at 4 °C for 5 min each wash. Bound proteins were released by two times 10-min incubation with 250 µL of 3xHA peptide (12149; Sigma) for DRM3 and 3xFLAG peptide (F4799; Sigma) for NRPE1 at room temperature. The eluted protein complexes were precipitated by trichloroacetic acid and subjected to mass spectrometric analyses as previously described (10).
Coimmunoprecipitation.
Approximately 1 g of flowers from each parental line, as well as F1 plants expressing complementing, epitope-tagged versions of both proteins, was grounded in liquid nitrogen with 5 mL of lysis buffer (LB), and the lysate was cleared by centrifugation at 10,000 × g in microfuge tubes for 10 min at 4 °C. The supernatants were incubated with 100 µL of either M2 Flag agarose (A2220; Sigma) or anti-HA affinity matrix (11815016001; Roche) for 2 h at 4 °C with rotation. The beads were then washed five times, for 5 min, with 1 mL of LB and resuspended in 100 µL of SDS/PAGE loading buffer. The various proteins were detected by Western blotting using either ANTI-FLAG M2 Monoclonal Antibody-Peroxidase Conjugate (A 8592; Sigma) at a dilution of 1:5,000, anti-HA high affinity (3F10) monoclonal antibody (11867431001; Roche) at a dilution of 1:3,000, anti-Myc 9E10 antibody (AFC-150P; Covance) at a dilution of 1:3,000, and endogenous AGO4 antibody at dilution of 1:1,000. All Westerns were developed using ECL Plus Western Blotting Detection System (RPN2132; GE Healthcare).
Genome-Wide ChIP Sequencing and Library Generation.
Two grams of tissue were ground in liquid nitrogen, and ChIP was performed as previously described (20) using endogenous NPRE1 antibody. As further confirmation, we performed ChIP twice from 2 g of plant extracts expressing NRPE1-FLAG either in WT or drm3 mutant background by FLAG beads. ChIP-enriched DNAs from the three biological replicates were subjected to library preparation and sequencing following the manufacturer’s instructions (Illumina). Reads were mapped to the TAIR10 genome using Bowtie (42) by allowing up to one mismatch and keeping only reads that uniquely map to the genome. Reads mapping to identical locations were collapsed into one read. Pol V sites were called using MACS (44) with a P value cutoff of 1e−1, using the WT ChIP-seq library as the treatment and the nrpe1 library as the control library.
Pol V Modeling.
To model Pol V occupancy at Pol V sites, we used methylation data derived from the whole-genome bisulfite libraries and small RNA data from the libraries described above. Cytosine abundance was derived from the TAIR10 build of the Arabidopsis genome. Modules were built using the random Forest package (45) using the default parameters. To test the performance of model prediction, the model was trained with ∼75% of the identified 4317 Pol V peaks and then tested with the remaining 25% of peaks. This training/testing was iterated 25× for each genotype and subsequent variable subtraction experiments. The reported distribution of R2 values is based on the correlation between the actual Pol V occupancy and predicted Pol V occupancy for the test sites of each iteration. For prediction of Pol occupancy at mutant loss/gain sites, all of the Pol V sites were used in model training, with the exception of those sites classified as affected in the mutant to be tested (i.e., when using drm3 data to train the model to predict Pol V occupancy at drm2 loss/no change/gain sites, those sites that experience loss/gain of Pol V in drm2 were not included in the drm3 training data) to avoid overfitting of the model.
Supplementary Material
Acknowledgments
We thank the staff members at the University of California, Los Angeles Broad Stem Cell Research Center BioSequencing core for high-throughput sequencing and Craig Pikaard for Pol V and AGO4 antibodies. C.J.H. is a Howard Hughes Medical Institute Fellow of the Damon Runyon Cancer Research Foundation. M.G. is supported by European Molecular Biology Organization Long-Term Fellowship ALTF986-2011. This work is supported by the US Department of Agriculture National Institute of Food and Agriculture (Hatch 1002874) (to X.Z.), NIH Grant GM089778 (to J.A.W.), and NIH Grant GM60398 (to S.E.J.). S.E.J. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE61192).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423603112/-/DCSupplemental.
References
- 1.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293(5532):1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 2.Beard C, Li E, Jaenisch R. Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev. 1995;9(19):2325–2334. doi: 10.1101/gad.9.19.2325. [DOI] [PubMed] [Google Scholar]
- 3.Fedoroff NV. Presidential address: Transposable elements, epigenetics, and genome evolution. Science. 2012;338(6108):758–767. doi: 10.1126/science.338.6108.758. [DOI] [PubMed] [Google Scholar]
- 4.Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6(5):351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- 5.Martienssen RA, Colot V. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science. 2001;293(5532):1070–1074. doi: 10.1126/science.293.5532.1070. [DOI] [PubMed] [Google Scholar]
- 6.Finnegan EJ, Dennis ES. Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res. 1993;21(10):2383–2388. doi: 10.1093/nar/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saze H, Mittelsten Scheid O, Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet. 2003;34(1):65–69. doi: 10.1038/ng1138. [DOI] [PubMed] [Google Scholar]
- 8.Kishimoto N, et al. Site specificity of the Arabidopsis METI DNA methyltransferase demonstrated through hypermethylation of the superman locus. Plant Mol Biol. 2001;46(2):171–183. doi: 10.1023/a:1010636222327. [DOI] [PubMed] [Google Scholar]
- 9.Cao X, Jacobsen SE. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol. 2002;12(13):1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- 10.Du J, et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell. 2012;151(1):167–180. doi: 10.1016/j.cell.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindroth AM, et al. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292(5524):2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 12.Zemach A, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153(1):193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stroud H, et al. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol. 2014;21(1):64–72. doi: 10.1038/nsmb.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan SW, et al. RNA silencing genes control de novo DNA methylation. Science. 2004;303(5662):1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- 15.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299(5607):716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 16.Kanno T, et al. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet. 2005;37(7):761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- 17.Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 2009;41(5):630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135(4):635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Law JA, et al. A protein complex required for polymerase V transcripts and RNA-directed DNA methylation in Arabidopsis. Curr Biol. 2010;20(10):951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong X, et al. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat Struct Mol Biol. 2012;19(9):870–875. doi: 10.1038/nsmb.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson LM, et al. SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature. 2014;507(7490):124–128. doi: 10.1038/nature12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu ZW, et al. The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS Genet. 2014;10(1):e1003948. doi: 10.1371/journal.pgen.1003948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong X, et al. Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell. 2014;157(5):1050–1060. doi: 10.1016/j.cell.2014.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henderson IR, et al. The de novo cytosine methyltransferase DRM2 requires intact UBA domains and a catalytically mutated paralog DRM3 during RNA-directed DNA methylation in Arabidopsis thaliana. PLoS Genet. 2010;6(10):e1001182. doi: 10.1371/journal.pgen.1001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152(1-2):352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431(7004):96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 27.Gowher H, Liebert K, Hermann A, Xu G, Jeltsch A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J Biol Chem. 2005;280(14):13341–13348. doi: 10.1074/jbc.M413412200. [DOI] [PubMed] [Google Scholar]
- 28.Kareta MS, Botello ZM, Ennis JJ, Chou C, Chédin F. Reconstitution and mechanism of the stimulation of de novo methylation by human DNMT3L. J Biol Chem. 2006;281(36):25893–25902. doi: 10.1074/jbc.M603140200. [DOI] [PubMed] [Google Scholar]
- 29.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J Biol Chem. 2004;279(26):27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 30.Ooi SK, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448(7154):714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hashimoto H, Vertino PM, Cheng X. Molecular coupling of DNA methylation and histone methylation. Epigenomics. 2010;2(5):657–669. doi: 10.2217/epi.10.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng B, et al. Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 2009;23(24):2850–2860. doi: 10.1101/gad.1868009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Law JA, et al. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature. 2013;498(7454):385–389. doi: 10.1038/nature12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang L, et al. An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat Struct Mol Biol. 2009;16(1):91–93. doi: 10.1038/nsmb.1539. [DOI] [PubMed] [Google Scholar]
- 35.El-Shami M, et al. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007;21(20):2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ream TS, et al. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell. 2009;33(2):192–203. doi: 10.1016/j.molcel.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haag JR, et al. In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Mol Cell. 2012;48(5):811–818. doi: 10.1016/j.molcel.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greenberg MV, et al. Interplay between active chromatin marks and RNA-directed DNA methylation in Arabidopsis thaliana. PLoS Genet. 2013;9(11):e1003946. doi: 10.1371/journal.pgen.1003946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xi Y, Li W. BSMAP: Whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu C, Meyers BC, Green PJ. Construction of small RNA cDNA libraries for deep sequencing. Methods. 2007;43(2):110–117. doi: 10.1016/j.ymeth.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 41.Henderson IR, et al. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet. 2006;38(6):721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- 42.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002;2(3):18–22. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.