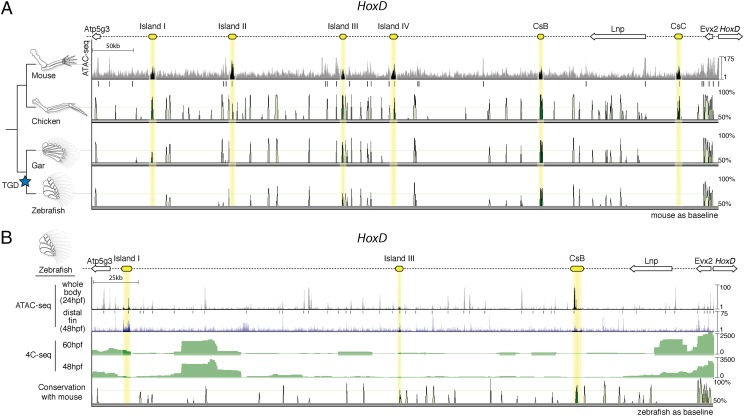
Fig. 1.
Chromatin state and sequence conservation of the HoxD autopod “regulatory archipelago” gene desert among select vertebrates. (A) (Top) Schematic representation of the HoxD centromeric gene desert, with cis-regulatory “islands” active in mouse denoted in yellow. ATAC-seq data are shown for mouse autopods at e12.5, providing a view of open chromatin. Statistically significant peaks are denoted by black bars. Sequence conservation is shown below for chicken, gar, and zebrafish. Note that sequence conservation for Island I is only found in gar, a nonteleost actinopterygian, and not in the teleost zebrafish. A blue star marks the teleost-specific genome duplication (TGD). (B) The zebrafish hoxD regulatory archipelago, with candidate “autopod” enhancers shown in yellow. ATAC-seq results for 24 hpf whole-body and 48 hpf distal fin are shown, identifying areas of open chromatin. 4C-seq results on whole-body 48 and 60 hpf embryos using hoxd13a as the target are shown in green. The putative teleost ortholog of Island I shows significant interaction with the hoxd13a promoter at 60 hpf. Vista plot with zebrafish as the baseline shows no sequence conservation with mouse for autopod enhancers other than Island III and CsB.