Fig 2. Proteolytic and photonic activation of plasminogen.
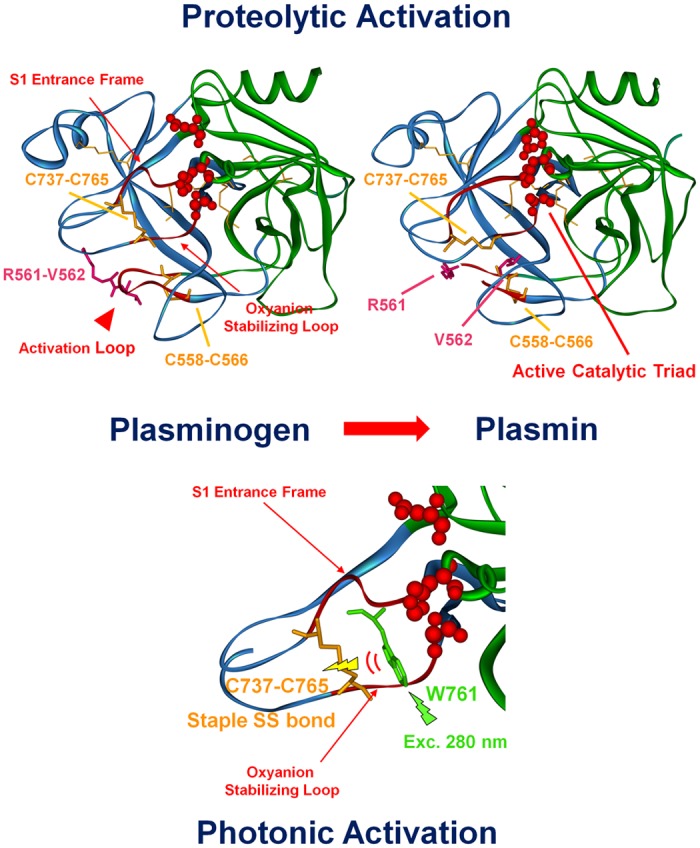
3D structures of the catalytic serine protease domain in plasminogen (to the left) and plasmin (to the right) (Ala543-Asn791). The xyz atomic coordinates for structural display were extracted from the crystallized structures of Glu1-Pmg (4A5T.pdb) [41] and activated plasminogen upon tight association with streptokinase (1BML.pdb) [4]. The C and N subdomains of plasminogen are displayed in blue and green, respectively. The disulphide bonds are displayed in stick configuration (orange) and two important disulphide bonds are highlighted: Cys556-Cys566 and Cys737-Cys765. The Trp and Tyr residues in the Serine Protease domain are displayed in line configuration (dark green). The active site residues (catalytic triad His603, Asp646 and Ser741 for plasminogen, His603, Asp646 and mutated Ala741 for plasmin) are displayed in CPK red (0.5 Å radius). The three important loops involved in plasminogen activation are displayed as red CPK: the oxyanion stabilizing loop, the S1 entrance frame and the activation loop. Top panel: The peptide bond Arg561-Val562 (in pink) in the activation loop is cleaved upon streptokinase activation leading to a conformational change and activating the catalytic triad (plasmin image to the right). Bottom panel: we can see in more detail the region around the oxyanion stabilizing loop, the S1 entrance frame. We observe a conformational change upon plasminogen activation involving the disulphide bond Cys737-Cys765 and Trp 761. We assume that upon UV illumination of plasminogen, Trp761 transfers its excitation energy to Cys737-Cys765. This results in disruption of this disulphide bond, leading to a conformational change that renders the catalytic triad active.
