Abstract
New Delhi metallo β-lactamases are one of the most significant emerging resistance determinants towards carbapenem drugs. Their persistence and adaptability often depends on their genetic environment and linkage. This study reports a unique and novel arrangement of bla NDM-1 gene within clinical isolates of Pseudomonas aeruginosa from a tertiary referral hospital in north India. Three NDM positive clonally unrelated clinical isolates of P. aeruginosa were recovered from hospital patients. Association of integron with bla NDM-1 and presence of gene cassettes were assessed by PCR. Genetic linkage of NDM gene with ISAba125 was determined and in negative cases linkage in upstream region was mapped by inverse PCR. In which only one isolate’s NDM gene was linked with ISAba125 for mobility, while other two reveals new genetic arrangement and found to be inserted within DNA directed RNA polymerase gene of the host genome detected by inverse PCR followed by sequencing analysis. In continuation significance of this novel linkage was further analyzed wherein promoter site detected by Softberry BPROM software and activity were assessed by cloning succeeding semi-quantitative RT-PCR indicating the higher expression level of NDM gene. This study concluded out that the unique genetic makeup of NDM gene with DNA-dependent-RNA-polymerase favours adaptability to the host in hospital environment against huge antibiotic pressure.
Introduction
The New Delhi Metallo-β-lactamase (NDM) has emerged as a major carbapenemase with rapid dissemination worldwide [1]. Clinical isolates harbouring NDM gene are often referred as superbugs. The NDM gene has been identified usually in enterobacteriaceae and recently in Pseudomonas spp. [2] which expresses resistance towards carbapenems and represents a significant threat for clinicians. This situation has been further complicated by the association of NDM gene with other resistant determinants [3]. Most importantly, however they represent an important stage in the evolution of antibiotic era.
Transfer of the bla NDM among promiscuous plasmids is the major dissemination route including clonal outbreaks. Noticeably, bla NDM has been frequently identified from unrelated gram negative bacilli, harboured by different plasmid types [4]. However, the mechanism of multiresistance trait of NDM positive isolates remained wanted and speculated that it could have been captured from original chromosomal location by mobile genetic elements. It has been reported that the bla NDM can be carried by different plasmid types (IncA/C, IncF, IncL/M, or untypable) and occasionally found to be chromosomally integrated [5]. The sequence of few plasmids carrying bla NDM are now available and reveals its inimitable genetic features such as association with insertion sequences and transposons for high mobility, acquisition of other multiresistance regions (aadB, dfrA12, bla OXA-30, aacA4) including additional important enzymes which make it more versatile [6].
In this study we have presented a unique genetic makeup of bla NDM gene among clinical isolates along with other resistance factors. However, this novel finding suggests the insertion of bla NDM within DNA directed RNA polymerase gene, thus favouring its survivability within hospital.
Materials and Methods
Bacterial isolates & Carbapenem susceptibility
A total of 105 consecutive, non-duplicates, carbapenem non-susceptibile isolates of P.aeruginosa were collected from indoor patients of Sir SunderLal Hospital, Banaras Hindu University, Varanasi, India, during March 2011 to September 2011. Identification of organisms was done by the conventional methods [7].
The metallo β-lactamase status of the strains was established by the Imipenem—Ethylene diamine tetra-acetic acid (EDTA) disc potentiation method [8]. A previously confirmed and sequenced clinical isolate of E. coli harbouring bla NDM was taken as positive control [9] and E. coli ATCC 25922 was used as negative control.
All MBL positive isolates were suspended in 1mL of buffered peptone water supplemented with 30% glycerol (peptone glycerol) and were kept at −80°C. The isolates were also stored in minimal media (10% Peptone, 5% Sodium Chloride and 0.8% Agar) as stab culture. ERIC-PCR was performed for genotyping by using their respective primers to determine clonal relatedness of the isolates [10].
Ethics Statement
This work and obtained samples specifically for this study has been ethically approved by the chairperson of the ethical committee of Institute of Medical Sciences, Banaras Hindu University, Varanasi; Ref. No.-Dean/2012-13/114.
Genotypic detection of blaNDM and their association with mobile genetic elements
Genomic DNA for PCR was extracted by using QIAamp DNA mini kit (Qiagen, Germany). Genotypic detection of bla NDM was performed by PCR assay in all the MBL positive isolates. Reaction conditions and primers used for PCR amplification were as described earlier [11]. Consecutively, the presence of integrons were examined by PCR, using primers to amplify a 160 bp fragment for class 1 integrase, and 288 bp fragment for class 2 integrase. The primers, PCR conditions and reaction mixtures used were as described previously [12]. To find the genetic association of bla NDM gene with integrons, PCR was performed using forward primer of the conserved region (5’CS) of integron gene and reverse primer of the characterized bla NDM gene [12]. Integrons contain multiresistant regions known as gene cassettes which were determined by 59 base elements PCR (Table 1) [13].
Table 1. List of primers used in the study.
| Primers | Target | Sequence (5’–3’) | Reference |
|---|---|---|---|
| HS287 F HS286 R | 59be association | GGGATCCGCSGCTKANCTCVRRCGTTAGSC GGGATCCTCSGCTKGARCGAMTTGTTAGVC | 12 |
| Inv 1F Inv 1R | To amplify flanking region | ATGGAAACTGGCGACCAACGG AATCGTCGGGCGGATTTCACC | Present study |
| POL’F POL’R | To amplify flanking region | ACGTCAGAGCGATGAAGACG GACCTGGAACTGACCGTACG | Present study |
| NDM 3P | 3’ end of linkage with DNA directed RNA Polymerase reverse | GATCGTGATGAGCCATTCCGCC | Present study |
| Native Pm Mutated Pm NDMc | To amplify RNA polymerase promoter along with integrated NDM gene | AACCTGATTGTCGAGCTCTACTC CTCGCTCTACTCCAAGTAAGAAC* CATCGAAATCGCGCGATGGCAG | Present study |
*underlined regions are mutated bases
Insertion sequences (IS), one of the major genetic elements in transposition of antibiotic resistance genes. In order to assess the linkage of NDM gene with the insertion sequence in our study, PCR analysis was performed by using forward primer of ISAba125 (Acinetobacter specific), and reverse primer of NDM gene. The reaction mixture and running conditions were as described earlier [14].
Detection of Novel linkage by inverse PCR
In order to determine the genetic environment surrounding the bla NDM-1 gene (apart from their linkage with ISAba125 and integrons) inverse PCR was performed. DNA was extracted from the three strains and digested with Sau3AI (Biolabs, US). DNA fragments obtained were then autoligated at 16°C with T4 DNA ligase in higher dilution to allow self circularization. The fragment of DNA containing the bla NDM-1 gene was used as a template for an inverse PCR with primers designed from the bla NDM-1 gene sequence. Inv1F as forward and Inv1R as reverse primer [Table 1] were used for amplification. Reaction mixture was approximately 5 ng of template DNA, 10 pmol of each primer, 200 μM deoxynucleoside triphosphate (dNTP) mix, 2 mM MgCl2 and of Taq DNA polymerase in the reaction buffer supplied with the enzyme. Reaction conditions were: initial denaturation at 94°C for 3 min followed by 32 cycles at 94°C for 30 s, 58°C for 45 s, 72°C for 1 min 25 s, and final extension at 72°C for 8 min.
Once the sequence of amplified product of inverse PCR was obtained, the linkage was confirmed by using POL’F 5′ACGTCAGAGCGATGAAGACG-3′ as forward primer and Inv1R 5′AATCGTCGGGCGGATTTCACC-3′ as reverse primer to detect 5’ region of target site, followed by second PCR reaction with NDM 3PF 5′GATCGTGATGAGCCATTCCGCC3′ as forward primer and POL’R 5′GACCTGGAACTGACCGTACG3′ as a reverse primer to confirm 3’ target region. To, further establish this linkage, the PCR reaction was performed using POL’F as forward and POL’R as reverse primer among NDM positive isolates.
Southern Hybridization
Simultaneously, to validate our study Southern blotting was performed on agarose gel by in-gel hybridization [15] with the bla NDM probe labelled with DIG HIGH PRIME LABELING MIX (ROCHE, Germany) detection Kit. The digoxigenin-labeled bla NDM specific probe was prepared using primers (Forward NDM 5′GGGCAGTCGCTTCCAACGGT 3′ and Reverse 5′CGACCGGCAGGTTGATCTCC 3′) that amplify a 130 bp region of the bla NDM gene. Total genomic DNA was digested by HindIII for fragmentation of DNA which is followed by the transfer to nylon membrane (Hybond N, Amersham, UK) and then hybridised with prepared bla NDM specific probe. Detection was performed by using an NBT color detection kit. (ROCHE,Germany). Promoter activity was assessed by designing two sets of primers; in first set forward primer (RNAPROM) was designed from upstream region of the RNA polymerase promoter whereas in the second set the primer with desired mutation was generated (MUTPROM) in the promoter region. For both the sets the reverse primer used was NDMc [Table 1]. The PCR amplicons were sequenced to confirm and cloned using pGEM-T vector (Promega, Germany) in E. coli JM107. Further, RNA was isolated from both the constructs using RNeasyR Mini Kit (Qiagen Hilden, Germany) and cDNA was prepared using Quanti Tect Reverse Transcription kit (Qiagen, Hilden, Germany). Thereafter, Semi quantitative reverse transcriptase PCR was performed to determine the expression of bla NDM in both the constructs.
Transferability & plasmid profiling
Horizontal transferability of bla NDM was investigated by transformation assay. Conjugation experiment was carried out between clinical isolates as donors and streptomycin resistance E.coli recipient strain B (Genei, Banglore, India). Overnight culture of bacteria were diluted in Luria Bertani broth (Hi-Media, Mumbai, India) and was grown at 37°C till the O.D. of the recipient and donor culture reached 0.8–0.9 at A600. Donor and recipient cells were mixed at 1:5 donor-to-recipient ratios and transconjugants were selected on imipenem (0.25mg/L) + streptomycin (1000mg/L) agar plates. Transformation was carried out using E.coli JM107 as recipient. Transformants were further selected on imipenem (0.25mg/L) containing LB agar plates.
For the detection of incompatibility group type of plasmid in transformants with bla NDM as well as in bla NDM harbouring donor strains, PCR based replicon typing was carried out targeting 18 different replicon types [16]. Plasmid stability and fitness was also assessed by serial passage of bla NDM positive isolates on LB broth without any antibiotic pressure. After each passage the isolates were tested for the presence of bla NDM PCR assay.
Detection of other resistant determinants
To investigate the presence of other resistant determinants along with NDM, isolates were further tested for the co-existence of other MBL genes such as bla IMP and bla VIM, as well as ESBLs, AmpC and class D carbapenemase (OXA-48, OXA-58, OXA-23 and OXA-198) genes by multiplex PCR [17–23]. In addition, the study of efflux pump activity of the strains was phenotypically detected by double disc synergy test using meropenem and CCCP (carbonyl cyanide m-chlorophenylhydrazone) [24]. Consequently the expression of the Mex-efflux system was determined by quantitative real time PCR assay. PA01 strain was used as control [25].
Antibiotic susceptibility testing
Antimicrobial sensitivity testing was performed on Mueller-Hinton agar (Hi-Media, Mumbai, India) plates by Kirby Bauer disc diffusion method and interpreted as per CLSI recommendations [26]. The antibiotic tested were amikacin (30μg), gentamicin (10μg), netilmicin (30μg), tobramicin (10μg), ceftazidime (30μg), ciprofloxacin (5μg), imipenem (10μg), meropenem (10μg), piperacillin/tazobactum (100/10μg) and polymyxin B (300μg) (Hi-Media, Mumbai, India). MICs of all the isolates were determined by the agar dilution method against cefotaxime, ceftazidime, ceftriaxone (Hi-Media, Mumbai, India), cefepime (Alembic Ltd., Vadodara, India), aztreonam (Aristo Pharmaceuticals Ltd., Mumbai, India), imipenem (United Biotech, Solan, India), meropenem (AstraZeneca Pharmaceuticals Ltd., Bangalore, India), tigecycline (Taj Pharmaceuticals Ltd., Mumbai, India), polymyxin (Celon laboratories Ltd, Andhra Pradesh, India). Escherichia coli ATCC 25922 was used as a control.
Sequencing analysis
All the PCR amplicons were purified using the QIAquick Gel Extraction kit (QIAGEN Inc., Valencia, CA) and were subjected to DNA sequencing (Merck, Bangalore, India). Sequences were analyzed using the BLAST suite of programs. (http://www.ncbi.nlm.nih.gov/BLAST/). Further, promoter sites were also determined by using Soft Berry BPROM software (http://linux1.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfindb).
Results
Genetic context of bla NDM
Among the 105 isolates, 38 strains exhibited MBL activity by Imipenem EDTA disc potentiation method, of which three nonclonal isolates (PA6, PA38 and PA47) were found to harbour bla NDM-1 (Table 2) and were carrying class 1 integron too. While out of the 35 MBL positive but bla NDM negative isolates, 7 and 3 isolates were harbouring VIM and IMP gene respectively. Remaining phenotypically MBL positive isolates did not showed any amplification with the targeted MBL gene primers (IMP, VIM, NDM). Further, class 1 integron was detected among the 99/105 (94.28%) isolates. None of the study isolates were found carrying class 2 integrase. On performing the ERIC PCR for all the 105 isolates a total of 46 different clonal types were observed. However two non MBL strains were found to be clonal with PA47.
Table 2. Clinical details of the isolates harbouring NDM gene.
| ID no. | Isolate | Patient age/Sex | Clinical specimen | Ward/OPD/ICU | Current diagnosis | Carbapenem Susceptibility | Other susceptible antibiotics tested in vitro | Coproduction of Class C | NDM or other MBL genes |
|---|---|---|---|---|---|---|---|---|---|
| PA6 | P.aeruginosa | 24year/female | Tissue | Burn | sepsis | R | Polymyxin B | CMY-2 | NDM |
| PA38 | P.aeruginosa | 25 year/female | Central tip | FICU | VAP | R | Polymyxin B | ACT, CMY-2 | NDM |
| PA47 | P.aeruginosa | 29 year/female | Pus | Ortho | sepsis | R | Polymyxin B | DHA, ACT | NDM,IMP, VIM |
OPD, outpatient department; ICU, Intensive care unit; VAP, ventilator-associated pneumonia; FICU, Female intensive care unit R, resistant; Class C-Plasmid mediated AmpC β -lactamases.
However, among the three NDM harbouring isolates 59be PCR amplification and their sequencing revealed presence of dihydrofolate reductase (dhfr) and aminoglycoside acetyl transferase (aac(6’)) within the gene cassette. While analysing the upstream linkage of NDM gene with ISAba125, isolate PA6 formed a band of 850 bp thus showing the association between them which was confirmed by sequencing. However, in case of PA38 and PA47 inverse PCR result gave a novel finding that bla NDM gene got integrated within DNA dependent RNA polymerase of host genome. This was further confirmed by designing primers which could amplify both 5’ and 3’ region of the insert (Fig. 1). This insertion was further verified and supported by the fact that the amplified product of higher basepair with Pol’F and Pol’R could be observed in PA38 and PA47 while expected size of amplification was evident with PA6 (Figs. 2 and 3). It was also observed that the recombinant bla NDM used promoter region of DNA dependent RNA polymerase for its expression (Fig. 4). Further, Semi quantitative reverse transcriptase PCR also establishes DNA dependent RNA polymerase promoter activity for higher expression of NDM gene (Fig. 5).
Figure 1. Schematic representation of genetic arrangement of bla NDM in Pseudomonas aeruginosa (PA38 & PA47).
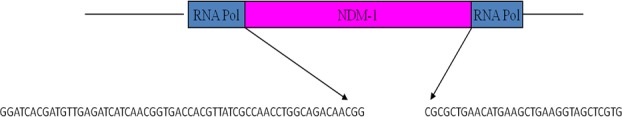
Section of DNA directed RNA polymerase gene in blue colored block while inserted NDM gene in pink block. NDM adjacent segments of DNA sequence were shown below.
Figure 2. Comparative analysis of PCR amplified product in different P. aeruginosa strains.
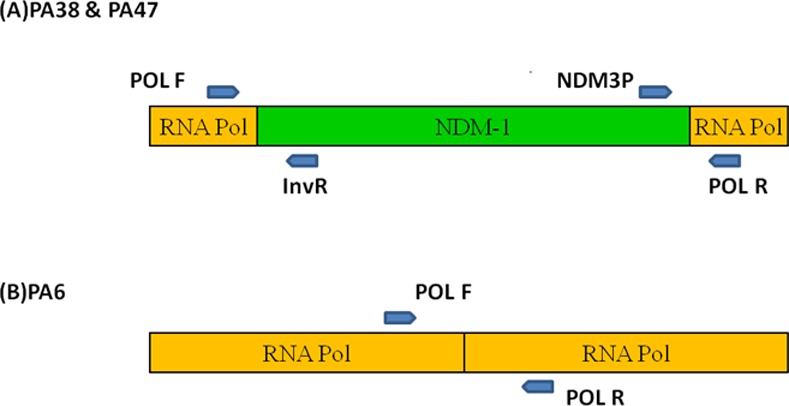
(A) NDM gene inserted in DNA directed RNA polymerase. (B) There is no insertion of NDM gene in RNA polymerase gene as intact gene segment of DNA directed RNA polymerase were detected through PCR mapping.
Figure 3. DNA directed RNA polymerase gene amplification in three NDM harbouring isolates.
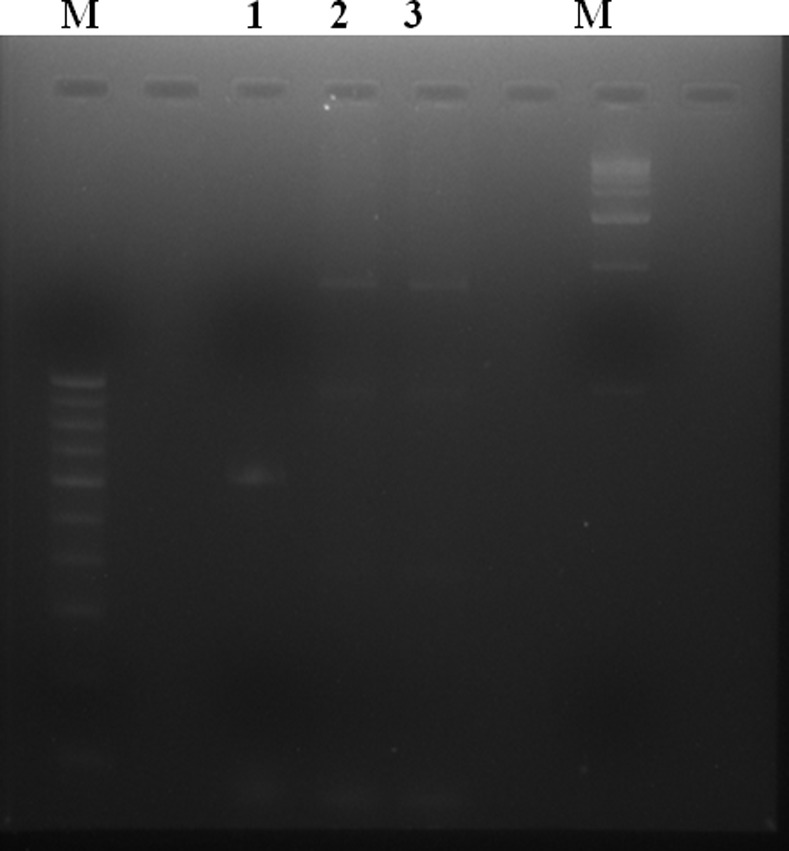
Lane 1- PA6 showing expected amplification of the intact gene; Lane 2 & 3- PA38 & 47 showing amplification of the gene along with the NDM insert (amplified product was comparatively of higher basepair than PA6).
Figure 4. Promoter sequence for NDM gene as determined by Softberry BPROM software.

(A) Promoter sequence of blaNDM within RNA polymerase gene in P. aeruginosa strain (AF047025) and (B) NDM promoter in A. baumannii associated with some insertion sequence (HQ857107). The −35 and −10 motifs of promoter are in boldface type whereas ORF sequence in red color font.
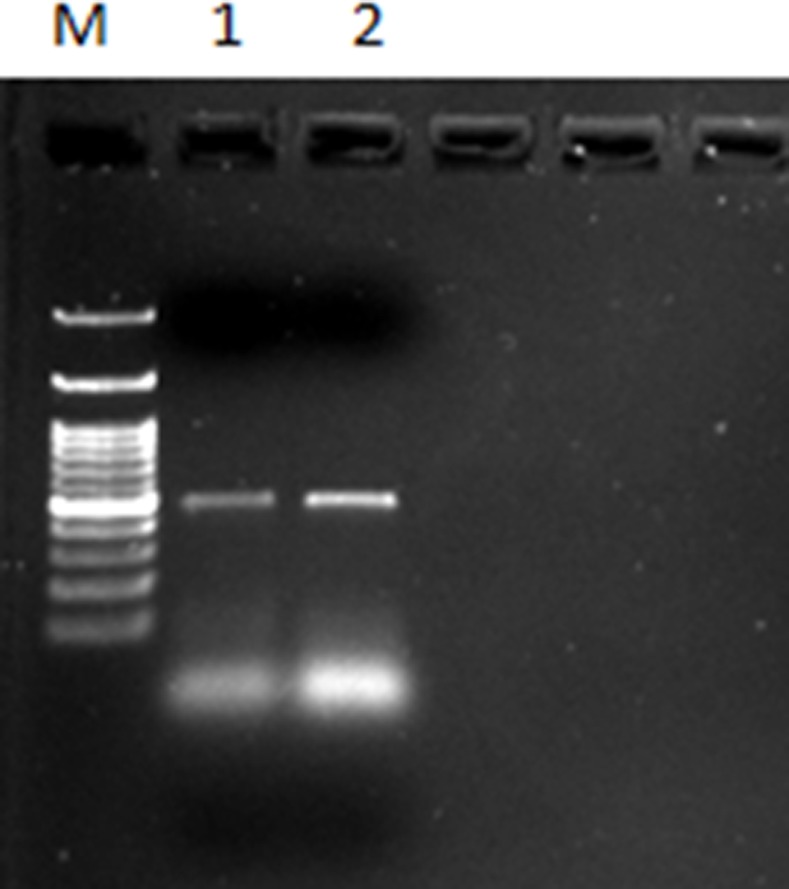
Figure 5. Semi quantitative reverse transcriptase PCR products showing promoter activity for expression of NDM gene, Lane1-NDM gene with mutated promoter (Low expression); Lane2- NDM with intact promoter showing higher expression.
Results of Southern hybridization were evident to suggest genetic background of the bla NDM-1. Plasmids of all three isolates were transferred to the nylon membrane while only PA6 hybridized with NDM-1 specific probe. Whereas when the same experiment was done with total DNA of all the test isolates, it was observed that all of them hybridized with the probe showing identical banding pattern with PA38 and PA47. Transformation and conjugation was successful only with PA6, where transconjugant showed to carry bla NDM within the 30–50kb plasmid. However, all the three isolates were carrying the plasmid of IncT type. The plasmid was highly unstable and was lost after six consecutive passages without any antibiotic in LB broth.
Coproduction of other resistant determinants
All the three isolates were negative for ESBLs but coproducing AmpC β-lactamase, while PA 47 was carrying bla IMP and bla VIM along with NDM gene. All of them were harbouring class D carbapenemase as well (Table 2). On performing the quantitative real time PCR to assess the level of transcription in MexAB-OprM efflux pump system, it was observed that the transcription level was 16-fold higher while under inducing condition when compared with PA01.
Antimicrobial susceptibility
On observing the antimicrobial susceptibility of these NDM producers, the isolates showed resistance towards all the tested antibiotics namely; cephalosporins, aztreonam, carbapenems, aminoglycoside and fluoroquinolones but were susceptible against polymyxin B. MIC of carbapenem was much higher for PA38 and PA47 compare to PA6 (Table 3).
Table 3. MIC (mg/L) of P. aeruginosa against cephalosporins and carbapenems.
| Isolate ID | CTX | CAZ | CEP | CFT | ERTA | IMP | MER |
|---|---|---|---|---|---|---|---|
| PA6 | ≥512 | ≥512 | ≥512 | ≥512 | 256 | 128 | 64 |
| PA38 | ≥512 | ≥512 | ≥512 | ≥512 | 512 | 512 | 512 |
| PA47 | ≥512 | ≥512 | ≥512 | ≥512 | 512 | 512 | 512 |
MIC, Minimum Inhibitory Concentration; CTX, Cefotaxime; CAZ, Ceftazidime; CEP, Cefepime; CFT Ceftriaxone; ERTA, Ertapenem; IMP,Imipenem; MER, Meropenem.
Discussion
Among the carbapenemases, the NDM gene has gained particular attention due to its global dissemination and multidrug resistance phenotype [27]. In the study, we found that all the three bla NDM harbouring strains were highly resistant to the antibiotics tested except polymyxin. Although the MBL enzymes donot affect monobactam, the co-existence of different AmpC genes such as bla EBC, bla DHA and bla CIT in these isolates were found to confer resistance to monobactam [14, 28]. Besides harbouring bla NDM the isolates were also carrying class D carbapenemases along with intrinsic mechanism. Similar genotype was earlier reported from India [29]. However, it leaves scope for future studies to address the exact role of six different carbapenem resistant determinants in a single isolate (PA47) when exposed to carbapenems.The study could demonstrate that bla NDM-1 in PA6 is plasmid mediated which was transferable by conjugation. However, same incompatibility type plasmid (lacking bla NDM-1) was present in other two isolates (PA38 and PA47). So, this plasmid could play the role of carrier for their acquisition of resistant determinant which later got integrated within the host genome.
In the previous study it was reported that the NDM gene has originated from Acinetobacter baumannii and is linked with ISAba125 in the upstream region. However, during horizontal transfer the gene got excised from the Acinetobacter DNA along with whole or truncated portion of ISAba125 [30]. Therefore Acinetobacter specific primer was used for the study and it was observed that only PA6 showed the presence of this insertion sequence in the upstream area. But acquisition of bla NDM within the DNA dependent RNA polymerase of host chromosome is quite unique and not reported previously.
The new genetic makeup of bla NDM under the control of RNA polymerase promoter might be responsible for higher level of expression, which could probably be the explanation for high MIC of PA38 and PA47.
The clinical challenge posed by bla NDM is currently higher worldwide due to its potential transferability. As our findings also suggest that the unique genetic makeup of bla NDM is for make an ease to endure the isolate within hospital environment to counter increasing antibiotic pressure. This is an important consideration that Pseudomonas with recombinant bla NDM is crucial for the hospital infection management and therapeutic options, otherwise this new genetic adaptability will lead in to more serious public health implications. From the study it cannot be truly predicted whether this recombination was a by chance event or a true event since the sample size was too small. Therefore, further investigation is advocated to establish this fact.
Acknowledgments
We are grateful to Head, Department of Microbiology, IMS and to staff of the S.S. Hospital, BHU for collecting the clinical isolates.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was financially supported by Department of Biotechnology, Govt. of India, New Delhi, India under the research project (P-07-467), Grant BT/PR11812/BRB/10/692. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Nordmann P, Poirel L, Toleman MA, Walsh TR, Livermore DM (2011) The emerging carbapenemases. Trends microbial 19: 588–595. 10.1016/j.tim.2011.09.005 [DOI] [PubMed] [Google Scholar]
- 2. Janvier F, Jeannot K, Tessé S, Nicoud MR, Delacour H, et al. (2013) Molecular characterization of blaNDM-1 in a ST235 Pseudomonas aeruginosa isolate, France. Antimicrob Agents Chemother 57: 3408–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dortet L, Nordmann P, Poirel L (2012) Association of the emerging carbapenemase NDM-1 with a bleomycin resistance protein in enterobacteriaceae and Acinetobacter baumannii , Antimicrob Agents Chemother 56: 1693–1697. 10.1128/AAC.05583-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Poirel L, Dortet L, Bernabeu S, Nordmann P (2011) Genetic feature of blaNDM-1 positive enterobacteriaceae. Antimicrob Agents Chemother 55: 5403–5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nordmann P, Dortet L, Poirel L (2012) Carbapenem resistance in Enterobacteriaceae: here is the storm. Trends Mol Med 18: 263–272. 10.1016/j.molmed.2012.03.003 [DOI] [PubMed] [Google Scholar]
- 6. Patridge SR, Iredell JR (2012) Genetic context of blaNDM-1. Antimicrob Agents Chemother 56: 6605–6607. [Google Scholar]
- 7. Collee JG, Miles RS, Watt B (1996) Tests for identification of bacteria. In Mackie & McCartney Practical Medical Microbiology (Edited by Collee JG, Marmion BP, Fraser AG, Simmons A). Churchill Livingstone, New York: 131–149. [Google Scholar]
- 8. Yong D, Lee K, Yum JH, Shin HB, Rossolini GM, et al. (2002) Imipenem–EDTA disk method for differentiation of metallo-β-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol 40: 3798–3801. 10.1128/JCM.40.10.3798-3801.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kumari S, Sen MR, Upadhyay S, Bhattacharjee A (2011) Dissemination of the New Delhi Metallo-β-lactamase-1 (NDM-1) among Enterobacteriaceae in a tertiary referral hospital in north India. J Antimicrob Chemother: 66(7):1646–47. 10.1093/jac/dkr180 [DOI] [PubMed] [Google Scholar]
- 10. Versalovic J, Koueth T, Lupski JR (1991) Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucl Acid Res 19: 6823–6831. 10.1093/nar/19.24.6823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yong D, Toleman MA, Giske CJ, Cho HS, Sundman K, et al. (2009) Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53: 5046–5054. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koeleman JGM, Stoof J, Vanderbij MW, Grauls V, Savelkoul P (2001) Identification of epidemic strains of Acinetobacter baumannii by Integrase Gene PCR. J Clin Microbiol 39: 8–13. 10.1128/JCM.39.1.8-13.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stokes HW, Holmes AJ, Nield BS, Nield BS, Holley MP, et al. (2001) Gene cassette PCR: Sequence-independent recovery of entire genes from environmental DNA. Appl Environ Microbiol 67:5240–5246. 10.1128/AEM.67.11.5240-5246.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mishra S, Sen MR, Upadhyay S, Bhattacharjee A (2013) Genetic linkage of blaNDM among nosocomial isolates of Acinetobacter baumannii from a tertiary referral hospital in north India. Int J Antimicrob Agents 41: 452–456. 10.1016/j.ijantimicag.2013.01.007 [DOI] [PubMed] [Google Scholar]
- 15. Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbour, N.Y: Cold Spring Harbour Laboratory Press; p32. [Google Scholar]
- 16. Carattoli A, Bertinia A, Villaa L, Falbo V, Hopkins KL, et al. (2005) Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63: 219–228. 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- 17. Senda K, Arakawa Y, Ichiyama S, Nakashima K, Ito H, et al. (1996) PCR detection of metallo-β-lactamase gene (blaIMP) in gram-negative rods resistant to broad spectrum β- lactams. J. Clin Microbiol 34: 2909–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tsakris A, Pournaras S, Woodford N, Palepou MF, Babini GS, et al. (2000) Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J Clin Microbiol 38: 1290–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Javier PF, Hanson ND (2002) Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex pcr. J Clin Microbiol 40:2153–2162. 10.1128/JCM.40.6.2153-2162.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bhattacharjee A, Sen MR, Prakash P, Anupurba S (2008) Role of beta-lactamase inhibitors in enterobacterial isolates producing extended-spectrum beta-lactamases.J Antimicrobl Chemother 61: 309–314. 10.1093/jac/dkm494 [DOI] [PubMed] [Google Scholar]
- 21. Jeong SH, Bae IK, Park KO, An YJ, Sohn SG, et al. (2006) Outbreaks of imipenem335 resistant Acinetobacter baumannii producing carbapenemases in Korea. J Microbiol 44: 423–431. [PubMed] [Google Scholar]
- 22. Dallenne C, Costa AD, Decre D, Favier C, Arlet G (2010) Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in enterobacteriaceae. J Antimicrob Chemother 65: 490–495. 10.1093/jac/dkp498 [DOI] [PubMed] [Google Scholar]
- 23. Poirel L, Marque S, He´ritier C, Segonds C, Chabanon G, et al. (2005) OXA-58, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii . Antimicrob Agents Chemother 49: 202–208. 10.1128/AAC.49.1.202-208.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Quale J, Bratu S, Landman D, Heddurshetti R (2003) Molecular Epidemiology and Mechanisms of Carbapenem Resistance in Acinetobacter baumannii Endemic in New York City. Clin Infect Dis 37: 214–220. 10.1086/375821 [DOI] [PubMed] [Google Scholar]
- 25. Mesaros N, Glupczynski Y, Avrain L, Caceres NE, Tulkens PM, et al. (2007) A combined phenotypic and genotypic method for the detection of Mex efflux pumps in Pseudomonas aeruginosa . J Antimicrob Chemother 59: 378–386. 10.1093/jac/dkl504 [DOI] [PubMed] [Google Scholar]
- 26. CLSI (2005) Performance standards for antimicrobial disc susceptibility test. CLSI: Wayne PA – M100-S15: [Google Scholar]
- 27. Kumaraswamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, et al. (2010) Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological and epidemiological study. Lancet Infect Dis 10: 597–602. 10.1016/S1473-3099(10)70143-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Berçot B, Poirel L, Dortet L, Nordmann P (2011) In vitro evaluvation of antibiotic synergy for NDM-1 producing enterobacteriaceae. J Antimicrob Chemother 66: 2295–2297. 10.1093/jac/dkr296 [DOI] [PubMed] [Google Scholar]
- 29. Kumarasamy K, Thirunarayan MA, Krishnan P (2010) Coexistence of blaOXA-23 with blaNDM-1 and armA in clinical isolates of Acinetobacter baumannii from India. J Antimicrob Chemother 65: 2253–2254. 10.1093/jac/dkq273 [DOI] [PubMed] [Google Scholar]
- 30. Toleman MA, Spencer J, Jones L, Walsh TR (2012) blaNDM-1 is a chimera likely constructed in Acinetobacter baumannii . Antimicrob Agents Chemother 56: 2773–2776. 10.1128/AAC.06297-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.


