Abstract
Objective
Vitamin D and the vitamin D receptor (VDR) appear to be important immunological regulators of inflammatory bowel diseases (IBD). Defective autophagy has also been implicated in IBD, where interestingly, polymorphisms of genes such as ATG16L1 have been associated with increased risk. Although vitamin D, the microbiome, and autophagy are all involved in pathogenesis of IBD, it remains unclear whether these processes are related or function independently.
Design
We investigated the effects and mechanisms of intestinal epithelial VDR in healthy and inflamed states using cell culture models, a conditional VDR knockout mouse model (VDRΔIEC), colitis models, and human samples.
Results
Absence of intestinal epithelial VDR affects microbial assemblage and increases susceptibility to dextran sulfate sodium-induced colitis. Intestinal epithelial VDR down-regulates expressions of ATG16L1 and lysozyme, and impairs anti-microbial function of Paneth cells. Gain and loss of function assays showed that VDR levels regulate ATG16L1 and lysozyme at the transcriptional and translational levels. Moreover, low levels of intestinal epithelial VDR correlated with reduced ATG16L1 and representation by intestinal Bacteroides in IBD patients. Administration of the butyrate (a fermentation product of gut microbes) increases intestinal VDR expression and suppresses inflammation in a colitis model.
Conclusions
Our study demonstrates fundamental relationship between VDR, autophagy, and gut microbial assemblage that is essential for maintaining intestinal homeostasis, but also in contributing to the pathophysiology of IBD. These insights can be leveraged to define therapeutic targets for restoring VDR expression and function.
Keywords: ATG16L1, Autophagy, bacteria, intestine, inflammation, nuclear receptor, transcriptional factor, vitamin D, Vitamin D receptor, Vitamin D response element (VDRE)
Introduction
Vitamin D deficiency is a critical factor in the pathology of IBD, colon cancer, autoimmune diseases, and other diseases.1-7 Low vitamin D status has been observed in IBD patients.8-10 Anorth-south gradient in rates of Crohn’s disease (CD) suggests that vitamin D deficiency may be an environmental trigger contributing to the pathogenesis of IBD.9 Vitamin D also influences the course and severity of IBD.9 The vitamin D receptor (VDR), a nuclear receptor,11 mediates most known functions of 1,25-dihydroxyvitamin D (Vitamin D3), the active form of vitamin D. VDR and vitamin D3 are recognized as key players in calcium homeostasis and in electrolyte and blood pressure regulation.12 More recent evidence demonstrates that vitamin D3, is an immunoregulatory hormone that modulates the innate and adaptive immune system.13-15 Vitamin D/VDR appears to be important immunological regulators of IBD.16, 17 Moreover, polymorphisms in the VDR gene are associated with increased susceptibility to IBD.18-22 VDR expression is significantly decreased in IBD patients.8, 23 Although it is possible that decreased VDR is a consequence of chronic inflammation in IBD, in experimental murine models of colitis, VDR knockout (KO) mice spontaneously develop colitis.24 Whole body VDR/IL-10 double KO mice develop a more severe colitis involving the entire intestinal tract.24 Mice lacking VDR activate the proinflammatory NF-κB pathway and are susceptible to Salmonella-colitis and chemical-induced colitis.25, 26 These findings suggest a critical role of VDR in maintaining intestinal homeostasis and possibly in causing/contributing to human IBD.
VDR also functions as a transcription factor.27 Target genes of VDR include anti-microbial peptide (AMPs) cathelicidin antimicrobial peptide 28, 29 (also called LL-37), β-defensin,29 and the 1, 25(OH) 2D3-regulated VDR-specific, Cyp24 hydroxylase gene. Indeed, ~3% of the mouse and human genomes are regulated directly or indirectly by the vitamin D endocrine system, further supporting the possibility of widespread effects of vitamin D and VDR in disease mechanisms.30-32 For example, epidemiological and experimental studies have indicated a protective action of vitamin D against colorectal cancer and upper GI cancers.33-40 Upregulation of VDR leads to induction of AMPs and the killing of intracellular Mycobacterium tuberculosis in human macrophages.41 Paneth cells are specialized intestinal epithelial cells located at the bottom of ileal crypts. The granules of Paneth cells contain AMPs—α-defensins, lysozyme, and secretory phospholipase A2.42-44 Paneth cells are a major source of monocyte chemotactic protein 1(MCP-1)45 and also produce the cytokine IL-17.46 A recent study demonstrated Paneth cells as a site of origin for intestinal inflammation.47 Thus, Paneth cells play a key role in innate immune responses and in shaping the gut microbiota.48 However, VDR regulation of Paneth cell function is not known.
Autophagy is a highly conserved process that is involved in intracellular homeostasis through the degradation and recycling of cytosolic contents and organelles, as well as in promoting the removal of intracellular microbes and immunity against infection.49, 50 Interestingly, three IBD susceptibility genes, IRGM, Nod2, and ATG16L1, are involved in autophagy.51-53 Deficits in the autophagy pathway can impair Paneth cell function in CD patients.54, 55 Autophagy can affect the biosynthesis or quality control of the lysosomal pathway in Paneth cell granules. Paneth cells with fewer granules might be a result of Paneth cell exhaustion, compensating for changes elsewhere in the epithelium.54 Expression in CD-like mutant Nod2 mouse alleles affect Paneth cells by reducing Nod2-dependent production of α-defensins in Paneth cells.56 Nod2 and ATG16L1 are in a common bacterial-sensing pathway that promotes bacterial antigen presentation. Nod2 recruits ATG16L1 to the plasma membrane at the bacterial entry site.57 Moreover, ATG16L1 may have unique protective functions, including AMP release in Paneth cells and the negative regulation of pro-inflammatory cytokine production.58, 59 Although vitamin D and autophagy are all involved in pathogenesis of IBD,60 it remains unclear whether these processes are related or function independently. Several studies61, 62 have identified vitamin D as a potent stimulator of autophagy in Mycobacterium tuberculosis infection and HIV infection. However, the crosstalk among VDR, autophagy, and bacteria in the gut remains unknown.
We have been investigating VDR63, 64 and bacterial inflammation.25, 63-66 We found that, on one hand, VDRs negatively regulate bacteria-induced NF-κB activity in intestinal inflammation.63 Lack of VDR leads to a reduction of IκBα, an endogenous inhibitor of NF-κB activity. On the other hand, bacteria regulate intestinal VDR expression in both gnotobiotic and bacterial-colitis models.63 Recent studies have also shown alternate bacterial profiles in VDR KO mice. VDR may regulate the gut microbes and probably contribute to maintenance of physiological host-microbe relationships. This could occur through several unique mechanisms that include NF-κB and autophagy. In the current study, we hypothesize that the intestinal epithelial VDR is a determinant of IBD risk through its actions on the autophagy gene (ATG16L1), thus determining states of Paneth cells and microbial assembly in intestinal homeostasis. We investigated mechanisms of intestinal epithelial VDR in healthy and inflamed states using a conditional knockout mouse model. We show that mice lacking VDR have increased bacterial loads in intestinal mucosa. The number of Paneth cells is decreased in the ileum of VDR−/− mice compared to control mice. We report that VDR levels correlated with levels of autophagy markers in vitro and in vivo. We further show that administration of the bacterial product butyrate increases intestinal VDR expression and suppresses inflammation in an experimental colitis model. Hence, our study fills an existing gap by characterizing the precise role of intestinal VDR in regulating intestinal homeostasis through changing the function of intestinal autophagy.
Materials and Methods
Human tissue samples
This study was performed in accordance with approval from the University of Rochester Ethics Committee (RSRB00037178). Colorectal tissue samples were obtained with informed consent from the sigmoid colon of 52 patients (51–83years old) exhibiting no apparent intestinal pathology and from the normal mucosa (i.e., >10cm colitis margin) of 30 patients undergoing anterior resection (44–85years old).
Animals
VDRloxP/loxP mice were originally developed by Dr. Geert Carmeliet.67 VDRΔIEC mice were obtained by crossing the VDRloxP/loxP mice with villin-cre mice (Jackson Laboratory, 004586). Experiments were performed on 2–3 months old mice. Mice were provided with water ad libitum, and maintained in a 12 h dark/light cycle. IL10−/− mice were purchase from Jackson Laboratory (002251). All work with animals were approved by the Rush University Animal Resources committee.
Induction of colitis
Mice were administered 5% DSS (MW = 40-50 kDa; USB Corp. Cleveland, OH) dissolved in filter-purified and sterilized water ad libitum for the experimental period. Animals were weighed daily. At day 7 after DSS administration, mice were sacrificed under anaesthesia. Severity of colitis was quantified by a disease activity index, determined by weight loss, fecal blood, and diarrhea.68
Butyrate treated mice model
Eight IL10−/− mice were divided into control and butyrate group. Butyrate group mice were given 2% sodium butyrate (Sigma, St. Louis, MO) water ad libitum for 3 weeks. The untreated control group was maintained on tap water throughout the experiment.
Co-housing experiment
Two to three month old female VDRloxP/loxP and VDRΔIEC were co-housed in new cages according to previously published methods.69 One cage contained 3 VDRloxP/loxP and 2 VDRΔIEC, another one contained 2 mice each. The mice were fed with the same food and water. After 4 weeks of co-housing, 5% DSS dissolved in filter-purified water was administered to the mice. Animals were weighed daily. At day 7 after DSS administration, mice were sacrificed under anesthesia.
Cell culture
Mouse embryonic fibroblasts (MEFs), human embryonic intestine INT 407 , HCT116 cells, and human colorectal adenocarcinoma SKCO-15 cells were grown in high glucose Dulbecco’s Modified Eagle Medium(DMEM) (Hyclone, SH30243.01) containing 10% (v/v) fetal bovine serum (GEMINI, 900-108), 50 μg/ml streptomycin, and 50 U/ml penicillin(Mediatech, Inc., 30-002CI). INT 407 cells were grown with 1% Non Essential Amino Acids (Mediatech, Inc., 25-025CI).
VDR knockdown of SKCO 15 with shRNA using cells retroviral GFP vector
SKCO-15 cells were transfected (Lipofectamine 2000, Invitrogen Corp.) with VDR shRNA retroviral GFP vector (OriGene, TG320568) plasmids targeting human VDR or control, scrambled shRNA on the same vector background (Origene) according to the manufacturer’s instructions. Briefly, SKCO-15 cells were seeded on day 1 in a six-well plate with complete medium and incubated overnight. Medium was replaced on day 2 with fresh complete medium containing 5 μg/ml puromycin dihydrochloride (GIBCO A11138-02) for 6 days. The transduced cells were sorted by GFP detection using flow cytometry and cultured in complete medium containing 10 μg/ml puromycin dihydrochloride for 13 passages.
Vitamin D-Responsive Element Transcriptional Activity
Cells were grown in triplicate and transfected with Cignal Vitamin D Reporter (luc) Kit (SABiosciences, Frederick, MD) using Surefect reagent (SABiosciences, Frederick, MD). The plasmid for the VDR reporter was a mixture of an inducible Vitamin D-responsive firefly luciferase construct and a constitutively expressing Renilla luciferase construct (40:1). The negative control was a mixture of a noninducible firefly luciferase construct and a constitutively expressing Renilla luciferase construct (40:1). After transfection for 24 hours, some cells were treated with 10 mM butyrate for 30 hours. Luciferase activity was determined using the Dual Luciferase Reporter Assay System (Promega) with a TD- 20/20 luminometer (Turner Designs, Sunnyvale, CA).
Western blot analysis and antibodies
Mouse ileal epithelial cells were collected by scraping from mouse ileum as previously described.65 Briefly, mouse epithelial cells were lysed in lysis buffer (1% Triton X-100 (Sigma-Aldrich, X100), 150 mM NaCl ( J.T.Baker 3624-19), 10 mM Tris ( Fisher Scientific, BP152-5) pH 7.4, 1 mM EDTA(Fisher Scientific, BP120-1), 1 mM EGTA(Sigma-Aldrich, E3889) pH 8.0, 0.2 mM sodium ortho-vanadate (Sigma-Aldrich, S6508), and protease inhibitor cocktail (Roche Diagnostics, 118367001)). Cultured cells were rinsed twice in ice-cold HBSS (Sigma-Aldrich, H1387), lysed in protein loading buffer (50 mM Tris, pH 6.8, 100 mM dithiothreitol (Amresco, 0281), 2% SDS (Sigma-Aldrich, L3771), 0.1% bromophenol blue (IBI Scientific, IB74040), and 10% glycerol (Sigma-Aldrich, G5516)), and sonicated (Branson Sonifier, 250). Equal amount of protein was separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose (Bio-rad, 162-0112), and immunoblotted with primary antibodies: VDR (Santa Cruz, sc13133), Beclin-1(Santa Cruz, sc10086), Villin (Santa Cruz, sc7672), LC3B (Cell Signal Technology Inc., 2775), ATG16L1 (Abgent, AP18176), p62 (Abgent, AP2183B) or β-actin (Sigma-Aldrich,A1978) antibodies and visualized by ECL chemiluminescence (Thermo Scientific, 32106).
Histology of Intestine
Intestines were harvested, fixed in 10% formalin (pH 7.4), processed, and paraffin embedded. Sections (5μm) were stained with H&E. For immunostaining, antigens were retrieved by 10-minute boiling in 10 mM citrate (pH 6.0). The slides were stained with antibodies as previously described.65 Blinded histological inflammatory scores were performed by a validated scoring system by a trained pathologist.70
Immunofluorescence
Ileal tissues from the distal portion of the ileum were freshly isolated and paraffin-embedded after fixation with 10% neutral buffered formalin. Immunofluorescence was performed on paraffin-embedded sections (5 μm). After preparation of the slides as described previously,71 tissue samples were incubated with anti-lysozyme (Santa Cruz, sc27958) at 4°C overnight. Samples were then incubated with sheep anti-goat Alexa Fluor 594 (Life technologies, A11058) and DAPI (Life technologies, D1306) for 1 hour at room temperature. Tissues were mounted with SlowFade (Life technologies, s2828), followed by a coverslip, and the edges were sealed to prevent drying. Specimens were examined with Zeiss laser scanning microscope (LSM) 710.
Fluorescence in situ hybridization
Fluorescent in situ hybridization (FISH) was performed using antisense ssDNA probes targeting the bacterial 16S rRNA. Bfra602 probe (5'- GAGCCGCAAACTTTCACAA -3') for Bacteroides fragilis group.72 Prior to performing the FISH assay, 5 μm tissue sections were baked over night at 55°C. Tissue sections were deparaffinized in xylene, dehydrated with 100% ethanol, air dried, incubated in 0.2M HCl for 20min and heated in 1 mM sodium thiocyanate at 80°C for 10 minutes. Samples were pepsin digested (4% pepsin in 0.01N HCl) for 20 minutes at 37°C, washed on slides in wash buffer (0.3 M NaCl, 0.03 M sodium citrate, pH 7, and 0. 1% SDS) and fixed on slides in 10% buffered formalin for 15 min, and hybridized with the probes at 5 ng/μl concentration each for 5 min at 96°C in hybridization buffer (0.9 M NaCl, 30% formamide, 20mMTris-HCl (pH 7.4), and 0.01% sodium dodecyl sulfate (SDS) and incubated at 37°C overnight. Slides were washed 4 times for 5 minutes each at 45°C in wash buffer. For visualization of the epithelial cell nuclei, the slides were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI)/ antifade solution. The slides were examined with Zeiss laser scanning microscope (LSM) 710.
Lysotracker staining
Lysotracker-red is a basic cell-permeable probe that accumulates in acidic vesicles. It is widely used to reflect lysosomal activity in live cells.61, 73, 74 Lysotracker staining was performed following the manufacturer protocol (Lonza Walkersville, Inc.). MEF and INT 407 cells were grown in the Lab-Tek Chambered Cover glass System (Thermo Scientific,154526), and the cells were then incubated with 100 nM LysoTracker Red lysosomal Probe(Lonza Walkersville,Inc., PA3015) in cell growth medium at 37°Cfor 60 min. After washing with HBBS, the cells were detected by fluorescence microscopy (AMG, EVOS fl).
Paneth cells counting
Paneth cells in mouse ileal cells were counted after anti-lysozyme immunofluorescence staining. The patterns of lysozyme expression in Paneth cells were classified into four categories: normal (D0), disordered (D1), depleted (D2) and diffuse (D3) according to published methods.59
Real Time quantitative PCR
Total RNA was extracted from mouse ileal epithelial cells or cultured cells using TRIzol reagent (Life technologies, 15596-02). RNA was first reverse-transcribed into cDNA with the iScript cDNA synthesis kit (Bio-Rad, 170-8891) according to the manufacturer’s manual. The RT-cDNA reaction products were subjected to quantitative real-time PCR using CTFX 96 Real-time system (Bio-Rad,C1000) and SYBR green supermix (Bio-Rad,172-5124) according to the manufacturer’s directions. All expression levels were normalized to β-actin levels of the same sample. Percent expression was calculated as the ratio of the normalized value of each sample to that of the corresponding untreated control cells. All real-time PCR reactions were performed in triplicate. Optimal primer sequences were designed using Primer-BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) or were obtained from Primer Bank (http://pga.mgh.harvard.edu/primerbank/) primer pairs listed Table 1
Table 1.
Real time PCR primers
| Gene name | Primers |
|---|---|
| mouse lyz1 F | GAGACCGAAGCACCGACTATG |
| mouse lyz1 R | CGGTTTTGACATTGTGTTCGC |
| mouse lyz2 F | ATGGAATGGCTGGCTACTATGG |
| mouse lyz2 R | ACCAGTATCGGCTATTGATCTGA |
| mouse DEFars1 F | AGCAGCCATTGTGCGAAGAA |
| mouse DEFars1 R | TGCTGTGTATTTGGAGCTTGG |
| mouse DEFa5 F | AGGCTGATCCTATCCACAAAACAG |
| mouse DEFa5 R | TGAAGAGCAGACCCTTCTTGGC |
| Mouse DEFa22-F | ACCAGGCTGTGTCTGTCTCCTT |
| Mouse DEFa22-R | TGGCCTCAGAGCTGATGGTTGT |
| Mouse RIP3g-F | GGTGAGGAGCATTAGTAACAGC |
| Mouse RIP3g-R | CCAGGGTTTAAGATGGTGGAGG |
| Mouse VDR-F | GAATGTGCCTCGGATCTGTGG |
| Mouse VDR-R | ATGCGGCAATCTCCATTGAAG |
| Mouse β-actin-F | TGTTACCAACTGGGACGACA |
| Mouse β-actin-R | CTGGGTCATCTTTTCACGGT |
Real-time PCR measurement of bacterial DNA
DNA was extracted from colonic tissues.75 16S rDNA PCR reactions were performed with the following primers: Universal bacteria 76 ( forward: 5’- TCCTACGGGAGGCAGCAGT -3’; reward:5’- GGACTACCAGGGTATCTAATCCTGTT -3’), E. Coli ( forward: 5’- CCTACGGGAGGCAGCAGT -3’; reward:5’-CGTTTACGGCGTGGACTAC -3’), Bacteroides fragilis (forward: 5’-GGCGCACGGGTG- AGTAACA-3’; reward:5’-CAATATTCCTCACTGCTGC-3’) and Butyrivibrio fibrisolvens (forward: 5’-CTAACACATGCAAGTCGAACG -3’; reward:5’-CCGTGTCTCAGTCCCAATG-3’), Primers specific to 18S rRNA77( forward: 5’-AGGGGAGAGCGGGTAAGAGA-3’; reward:5’-ggacaggactaggcg- gaaca-3’)were used as an endogenous control to normalize loading between samples. The relative amount of 16S rDNA in each sample was estimated using the ΔΔCT.
Mucosa microbial and fecal 454 Pyrosequencing
The tubes for microbial sampling were autoclaved and then irradiated with ultraviolet light to destroy the environmental bacterial DNA. The mice were then anesthetized and dissected. Fecal isolated freshly from the gut and placed into the specially prepared tubes, as described in our previously published papers.78, 79 The samples were kept at low temperature with dry ice and mailed to Research and Testing Laboratory, Lubbock, TX, for 454 pyrosequencing. The V4-V6 region of the samples was amplified in Research and Testing Laboratory, Lubbock, TX, for pyrosequencing using a forward and reverse fusion primer. The sequences were denoised, subjected to quality checking. Taxonomic identifications were assigned by queries against NCBI. Differences in microbial communities between VDRloxP/loxP and VDRΔIEC groups were analyzed, as we did in previous studies.78, 79Briefly, Principal Coordinates Analysis (PCoA) of unweighted UniFrac distances plots were plotted using quantitative insights into microbial ecology (QIIME).80 To determine differences in microbiota composition between the animal groups, the analysis of similarities (ANOSIM) function in the statistical software package PRIMER 6 (PRIMER-E Ltd., Lutton, UK) was used on the unweighted UniFrac distance matrixes.
Chromatin immunoprecipitation (CHIP) assay
Binding of VDR to the ATG16L1 promoter was investigated using the ChIP assay as described previously.25 Briefly, MEF cells were treated with 1% formaldehyde for 10 min at 37°C. Cells were washed twice in ice-cold phosphate buffered saline containing protease inhibitor cocktail tablets (Roche). Cells were scraped into conical tubes, pelleted and lysed in SDS Lysis Buffer. The lysate was sonicated to shear DNA into fragments of 200–1000 bp (4 cycles of 10 s sonication, 10 s pausing, Branson Sonifier 250, USA). The chromatin samples were pre-cleared with salmon sperm DNA–bovine serum albumin-sepharose beads, then incubated overnight at 4 °C with VDR antibody (Santa Cruz Biotechnology). Immune complexes were precipitated with salmon sperm DNA–bovine serum albumin-sepharose beads. DNA was prepared by treatment with proteinase K, extraction with phenol and chloroform, and ethanol precipitation. Searching mouse ATG16L1 gene, we found a similar sequence as the VDRE sequence “(G/A)G(G/T)TCA”. We then designed primers for ChIP. PCR was performed using the following promoter specific primers: ATG16L1 forward, 5’-GGTTCCGTTCTTGTTTCT- 3’; reverse, 5’-TCAAGTTGT- CTCCAAGATTAT- 3’.
Statistical Analysis
Data are expressed as mean± SD. Differences between two samples were analyzed by Student’s t test. Differences among three or more groups were analyzed using ANOVA with GraphPad Prism 5. P-values of 0.05 or less were considered statistically significant.
Results
Established intestinal VDRΔIEC model
We have established a VDRΔIEC model using the VDRloxP/loxP mouse strain.67, 81 As shown in figure 1A and 1B, there was no detectable VDR expression in intestinal epithelial cells of VDRΔIEC mice by immunohistochemistry or by Western blot analysis. These data confirm that the intestinal VDR knock out is established. The VDRΔIEC model allows us to: 1) focus on intestinal epithelial VDR and clearly define the mechanisms by which intestinal epithelial VDR regulates intestinal homeostasis; 2) study a well-controlled system without having to feed the mice a special calcium-enriched diet; and 3) avoid other non-digestive disorders found in total VDR−/− mice.
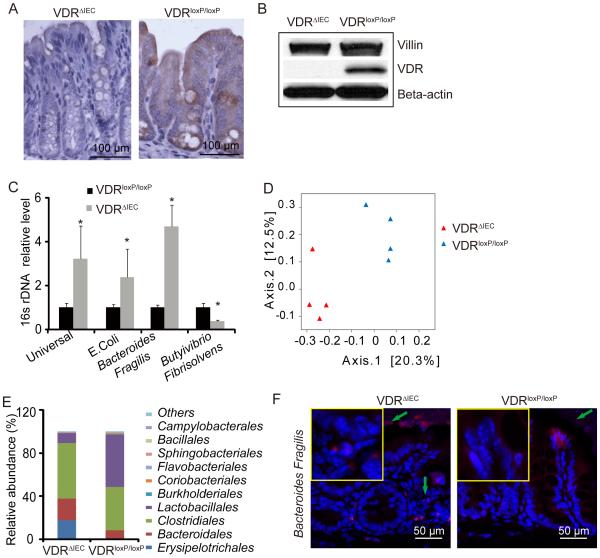
Figure 1.
Established intestinal epithelial cell VDR KO (VDRΔIEC) mice and their bacterial profile. (A) VDR immunohistochemistry staining in intestine. (B) Western blotting of intestinal VDR. Villin is a marker for IECs. (C) Real-time PCR of bacterial universal 16srDNA and 16s rDNA for E.coli, Bacteroides fragilis and, Butyrivibrio fibrisolvens. Primers specific to 18S rRNA were used as an endogenous control to normalize loading between samples. The relative amount of 16S rRNA in each sample was estimated using the ΔΔCT method. (n=3,* P<0.05) (D) Principal Coordinates Analysis (PCoA) of unweighted UniFrac distances of 16 S rRNA genes. Analysis for VDRloxP/loxP (blue) and VDRΔIEC (red) mice. The results indicate that fecal microbial communities differ in VDRΔIEC mice compared to VDRloxP/loxP. (E) Bacterial community of fecal samples from VDRΔIEC and VDRloxP/loxP mice, using 454 16S rRNA sequencing data.(n=4/group) (F) Representative fluorescence in situ hybridization staining for Bacteroides fragilis (red) in VDRΔIEC and VDRloxP/loxP control mice. Blue, DAPI.
Absence of intestinal epithelial VDR leads to ecologic change in bacterial profiles
Interestingly, bacterial abundance is significantly changed in VDRΔIEC mice compared with the VDRloxP/loxP mice without any treatment; demonstrating increased E. coli and Bacteroides and decreased butyrate-producing bacteria (Butyrivibrio) (figure 1C).Our 454 16S rRNA sequencing data also shown the altered bacterial profile in the VDRΔIEC mice. Principal Coordinate Analysis (PCoA) indicated that fecal microbial communities differ in VDRΔIEC mice compared to VDRloxP/loxP (figure1D). The relative abundance of bacteria was shifted in VDRΔIEC mice compared to VDRloxP/loxP (figure1E).These data are consistent with our 454 16S rRNA sequencing of cecal and proximal colon samples that revealed different taxa abundances with a significant increase in the proportion of Bacteroides in whole body VDR KO mice relative to WT mice (data not shown). These results are also consistent with a study which showed commensal Bacteroides species induce colitis.82 Bacteroides fragilis, a common human commensal microbiota, has been associated with IBD.83-85 To examine the distribution and abundance of bacteria, we performed FISH fluorescence in situ hybridization (FISH) staining for Bacteroides fragilis. Our data show enhanced Bacteroides fragilis in the VDRΔIEC mice compared with the VDRloxP/loxP mice (figure 1F).
VDRΔIEC mice are susceptible to chemical injury
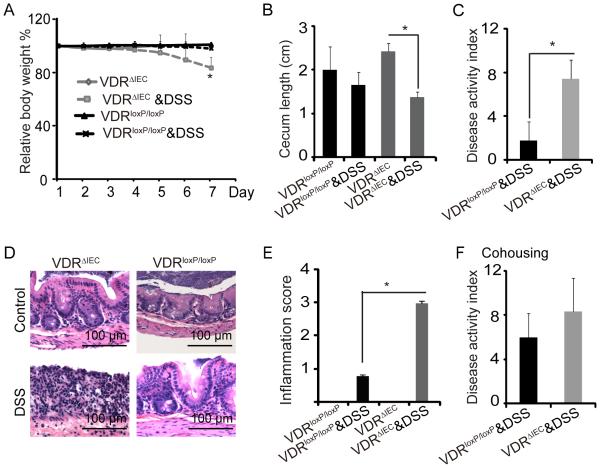
To investigate the biological effects of intestinal epithelial VDR in response to injury, we used a dextran sulfate sodium (DSS)-colitis model.86 Our data showed that VDRΔIEC mice were susceptible to DSS-mediated inflammation than VDRloxP/loxP mice (figure2). The VDRΔIEC mice had significant loss of body weight after DSS treatment for 7 days (figure 2A). The cecum length was significantly reduced in the VDRΔIEC mice with DSS compared to VDRΔIEC mice without DSS, whereas no significant change was found between VDRloxP/loxP mice with or without DSS (figure 2B). In VDRΔIEC mice with DSS, fecal blood was more obvious, stools were less formed, and more weight was lost. Accordingly, the Disease Activity Index was significantly increased compared to the VDRloxP/loxP group (P< 0.05). (figure. 2C) The H&E staining data in figure. 2D showed more severe intestinal inflammation in VDRΔIEC mice with DSS compared with the VDRloxP/loxP mice. Next, the level of intestinal inflammation was quantified histologically in a blinded fashion by a trained pathologist. The inflammation score in the DSS-treated VDRΔIEC mice was significantly higher than the DSS-treated VDRloxP/loxP mice (figure 2E). Since, the microbiota may play a role in the vulnerability of the VDRΔIEC mice to DSS, we examined the transmissibility of the phenotype by performing a co-housing experiment and then challenging the mice with DSS. We found that the co-housing increased disease activity index of VDRloxP-loxP mice to a level similar to that seen with the VDRΔIEC mice (figure 2F compared to 2C). The co-housing data suggest that the absence of intestinal epithelial VDR confers a transmissible risk for DSS-induced colitis. Taken together, our data indicate that intestinal VDR contributes to host protection against inflammation.
Figure 2.
VDRΔIEC mice have worse outcomes with DSS-induced colitis. (A) Relative body weight changes in mice with or without DSS. (B). Cecum shorten was found in VDRΔIEC mice with colitis. (C)Disease Activity Index of colon from mice with or without DSS treatment. (D) Representative H&E histology of colon from mice with or without DSS treatment. (E) Inflammatory scores of the mouse intestine with or without DSS. VDRloxP/loxP and VDRΔIEC mice were administrated with DSS. n=5/group, * P<0.05. (F) Inflammatory scores of intestine after co-housing VDRloxP/loxP and VDRΔIEC mice. Two to three month old female VDRloxP/loxP and VDRΔIEC were co-housed in new cages at 3:2 or 2:2 mice. After 4 weeks of co-housing, the mice were administered with 5% DSS for 7 days to induce colitis. The dysbiosis associated with VDR deficient mice appears to confer risk of chemically induced colitis. VDRloxP/loxP mice co-housed with VDRΔIEC mice developed the similar inflammation as the VDRΔIEC mice.
Absence of Intestinal epithelial VDR leads to abnormal Paneth cells
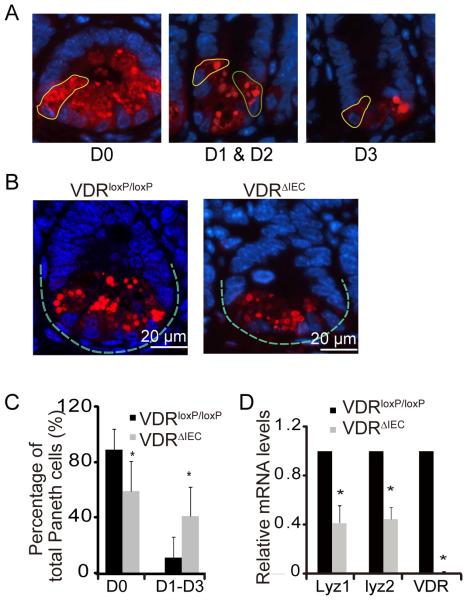
Paneth cells are specialized intestinal epithelial cells that secrete AMPs,42-44 sense commensal bacteria, and maintain homeostasis at the intestinal-microbial interface.48 We counted the number of Paneth cells, using a previously reported method to stain Lysozyme.59 Abnormal Paneth cells were grouped as D1 (disordered), 2 (depleted), and D3 (diffuse) (figure 3A). We found fewer than normal Paneth cells in the VDRΔIEC mice ileum than in VDRloxP/loxP mice (figure3B). The abnormal Paneth cells D1-D3 increased in the VDRΔIEC mice (figure 3C). Abnormal Paneth cells lead to decreased Lysozyme (Lyz1 and Lyz2) at the mRNA level (figure 3D).
Figure 3.
VDR affects patterns of Paneth cells in VDRΔIEC mice. (A) Representative images showed patterns of Paneth cells. D0, normal (yellow circle); D1, disordered (green circle); D2, depleted (yellow circle); D3, diffuse (yellow circle). (B) Representative images of indirect immunofluorescence of sections stained for lysozyme (red) in ileal crypts of VDRΔIEC and VDRloxP/loxP mice. (C) Percentage of Paneth cells displayed normal and abnormal (D1 to D3) patterns of lysozyme expression (n=10/group, * P<0.05). (D) Decreased lysozyme (lyz) in VDRΔIEC mice (n=3/group, * P<0.05).
Intestinal epithelial deletion of VDR down regulates ATG16L1 and autophagy that can impair Paneth cell function
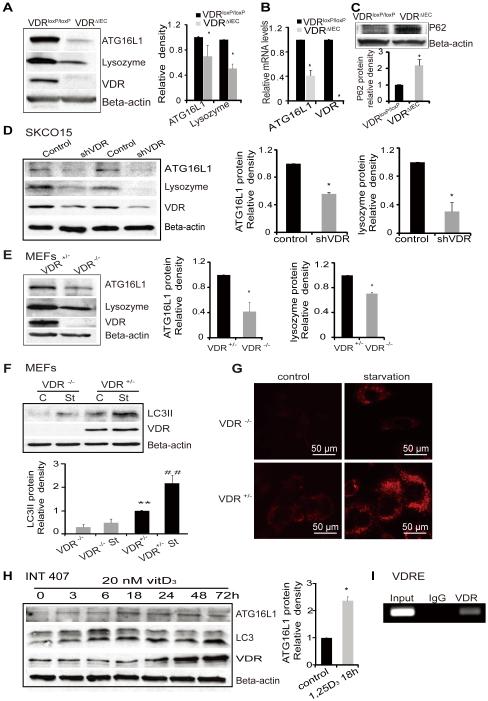
Deficits in the autophagy pathway can impair Paneth cell function. Autophagy plays an essential role in innate immunity.49 The IBD susceptibility geneATG16L1 is involved in autophagy and contributes to inflammation and dysbiosis.87 Our data showed that deletion of intestinal epithelial VDR leads to a significant reduction of ATG16L1 protein in vivo (figure4A).The protein level of lysozyme, another component of the autophagy pathway, was also significantly lower in VDRΔIEC mice (figure4A). We found that mRNA levels of ATG16L1 were lower in VDRΔIEC mice compared to the control VDRloxP/loxP mice (figure 4B). P62 is a signaling adaptor that accumulates in autophagy-deficient mice. 88, 89 We also found p62 protein increased in VDRΔIEC mice (figure 4C).
Figure 4.
VDR regulation of the expression levels of autophagy related genes. (A) Levels of ATG16L1 protein and lysozyme in VDRΔIEC and VDRloxP/loxP mice. The relative intensity of ATG16L1 and lysozyme. (n=3/group. *P<0.05). (B) ATG16L1 mRNA level decreased in VDRΔIEC (n=3, *P<0.05 ). (C) Levels of P62 protein in VDRΔIEC and VDRloxP/loxP mice. The relative intensity of P62. (n=3 /group. *P<0.05). (D) ATG16L1 and lysozyme were decreased in intestinal epithelial cells with knockdown VDR by SiRNA VDR. (E) Lacking of VDR induced the reductions of autophagy marks (ATG16L1 and lysozyme) in MEF cells. (F) Expression of LC3 II in VDR−/− and VDR+/− MEF cells. MEF cells were cultured to 80% confluence. Incubated in FBS free E-MEM for 2 hours as starvation (st). Cell lysates were immunoblotted with antibodies against LC3B and beta-actin. (G)Representative images of lysotracker staining in MEF VDR−/− and VDR+/− cells. MEF cells were grown in the Lab-Tek Chambered coverglass System, incubated in FBS free E-MEM for 2 hours as starvation, and incubated with 100 nM LysoTracker red lysosomal probe for 60 min. After washing with HBBS, the cells were detected under fluorescence microscopy (AMG, EVOS fl).(H) ATG16L1 was increased with the other autophagy markers (LC3, ATG16l1) in intestinal INT407 cells with enhanced VDR by vitamin D treatment. (I) Regions encompassing putative VDRE amplified by PCR in ChIP assays in vitro.
VDR knockout decreases ATG16L1 and LC3B proteins in SKCO15 and MEF cells
The ATG genes control autophagosome formation through ATG12-ATG5 and microtubule-associated protein1 light chain 3 (LC3-II or ATG8-II) complexes. In a loss of function experimental model, we further determined the effects of VDR on autophagy. We knocked down VDR protein with siRNA in human colon adenocarcinoma-derived epithelial SKCO15 cells. We found that VDR knockdown led to significantly decreased ATG16L1 and lysozyme at the protein levels (figure 4D).
We used VDR−/− and VDR+/− mouse embryonic fibroblast (MEF) cells to test whether one allele of the VDR gene was sufficient to re-establish a normal autophagy phenotype, and found that complete lack of VDR led to lower expression levels of ATG16L1, lysozyme, and LC3 II (figure 4E). Moreover, in a starvation-induced autophagy model, we found that the LC3 II level was significantly lower in VDR−/− MEF incubated in FBS free E-MEM medium, compared to VDR+/− after starvation, although LC3 II was increased by starvation in both VDR−/− and VDR+/− MEF cells (figure 4F). Using lysotracker Red to mark lysosome in vitro,90 we further showed differences in autophagy activity in VDR−/− and VDR+/− MEF cells (figure 4G). Without any treatment, the basal level of lysotracker Red uptake was already lower in VDR−/− MEF, compared to VDR+/− cells. After starvation for 2 hours to induce autophagy in cells, lysotracker Red uptake was enhanced. However, there was still less autophagy in the cells lacking VDR, compared to the VDR+/− MEF cells (figure 4G).
Enhanced VDR protein is associated with elevated autophagy markers within intestinal epithelial cells
Based on our observations, we further determined the possibility for restoration of intestinal epithelial VDR. We hypothesized that the status of VDR changes the expression of autophagy markers. Vitamin D3 is known to increase VDR by blocking ubiquitin/proteasome-mediated degradation.91 To explore the impact of enhanced VDR on autophagy, we investigated the response in the human colonic epithelial cell line INT 407. Vitamin D3 increased LC3 II/I associated with increased VDR in a time dependent manner. It also increased ATG16L1 associated with elevated level of VDR protein (figure 4H). Activated VDR is known to bind to the vitamin D-response element (VDRE) in the promoter of target genes to regulate gene transcription. We further identified the VDRE in the ATG16L1 gene by CHIP assay (figure 4I). Taken together, our data suggested that VDR expression status was able to change autophagy marker ATG16L1, LC3 II, and autophagy activity.
Decreased VDR expression is observed in UC patients and in an experimental colitis IL10−/− model
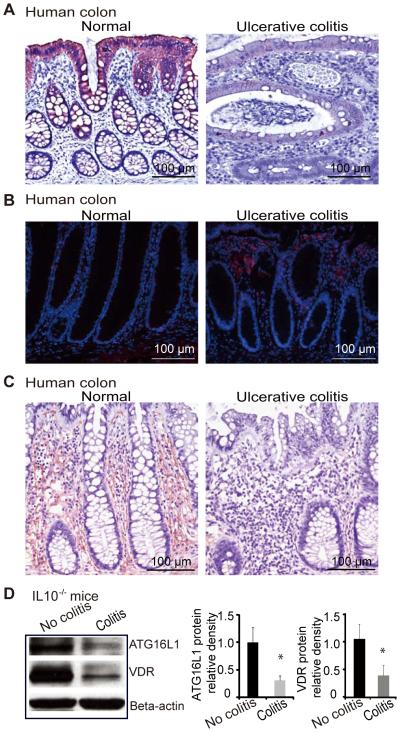
The VDR is associated with chronic inflammation.8, 23 Intestinal VDR expression is decreased in patients with UC as shown in figure 5A, which is consistent with the literature.8, 23 Moreover, in the inflamed human intestine with low VDR, increased Bacteroides was identified by FISH. Intestinal ATG16L1 staining is also very weak in patients with UC (figure 5C). We found a similar trend of VDR and ATG16L1 expression in an experimental colitis model, where IL10−/− mice spontaneously develop IBD.92 A significant decline in the expression of VDR was found in IL10−/− mice with colitis symptoms compared to mice without colitis symptoms (figure 5D). Moreover, we also found decreased ATG16L1 in intestinal epithelial cells in experimental colitis model (figure 5D).
Figure 5.
VDR expression in human intestine and colitis models. (A) Intestinal VDR staining is very weak in inflamed colon of Ulcerative colitis (UC) patients. (B) Representative FISH staining for Bacteroides fragilis (red) in tissues from UC patients. Blue, DAPI. Normal, normal tissue adjacent lesion area; Ulcerative colitis, lesion area. (C) Representative Intestinal ATG16L1 staining is very weak in inflamed colon of IBD patients. (D) Decreased VDR and ATG16L1 in the IL10−/− mice with colitis symptom. Data are presented as the mean ± SD from 5 mice/group.
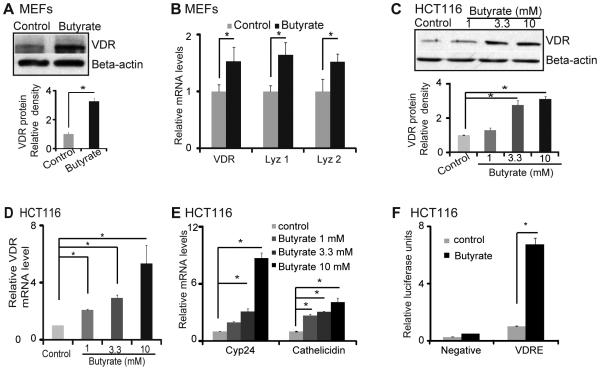
Bacterial product butyrate upregulates VDR in human epithelial cells in vitro
Our microbiome data (figure1C) indicate that butyrate-producing bacteria (Butyrivibrio) was decreased in the in VDRΔIEC mice. Moreover, a recent study indicate that dysbiosis in intestine leads to less butyrate-producing bacteria or less butyrate in intestine.93 We pretreated human intestinal epithelial cells with butyrate and found that it significantly increased VDR expression at the mRNA and protein levels (figure 6A and 6B). In the human intestinal epithelial cell line HCT116, VDR protein and mRNA were increased by butyrate in a dose–dependent manner (figure 6C and D). We further investigated VDR transcriptional activity following stimulation with butyrate in the human intestinal epithelial cell line HCT116. In cells transfected with the VDR reporter plasmid, butyrate significantly induced VDR transcriptional activity compared to control cells without butyrate stimulation. The relative fold increase was compared with cells transfected with the negative control plasmid (Negative) (figure 6E). Additionally, the expression of VDR target genes Cyp27 and cathelicidin increased significantly after butyrate treatment in the human epithelial cells (figure 6F).
Figure 6.
Bacterial product butyrate activates VDR signaling pathway in human intestinal epithelial cells. (A) Butyrate increased VDR protein level in MEF cells. (B) Butyrate increased VDR and lysozyme mRNA levels. (C) Butyrate increased VDR protein level in dose-dependent manner in HCT116 cells. (D) Butyrate increased VDR mRNA level in dose-dependent manner in HCT116 cells. (E) Butyrate increased the VDR transcription activity in human epithelial cells. HCT116 cells were transfected with Cignal Vitamin D Reporter (luc) Kit. After transfection for 24 hours, cells were treated with 10 mM butyrate for 30h, and luciferase activity was determined. Firefly luciferase activity was normalized to Renilla luciferase activity, and the activity was expressed as relative luminescence units. (F) Butyrate increased cyp24 and cathelicidin mRNA levels in dose-dependent manner.
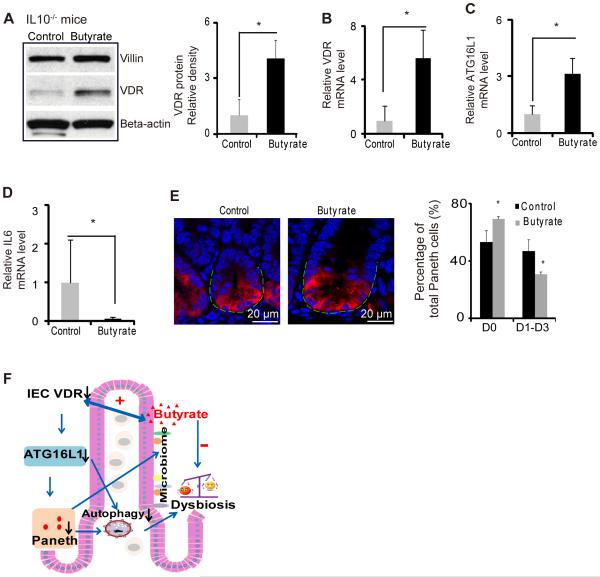
Butyrate treatment increases VDR and suppresses inflammatory cytokine IL-6 in a IL10−/− colitis model
We further used the experimental colitis IL10−/− model to investigate the in vivo physiological relevance of VDR regulation of epithelial function, including responses to bacterial products and anti-inflammatory effects. Butyrate treatment significantly increased VDR at the mRNA and protein levels in the IL10 knockout mice (figure 7A&B). After butyrate treatment, intestinal ATG16L1 was also significantly increased at the mRNA level (figure 7C) and suppression of inflammatory cytokine IL-6 was also observed in the IL10 knockout mice (figure 7D). Meanwhile, butyrate treatment increased the normal Paneth cells ratio (figure 7E). These data suggest that enhancing VDR expression by a bacterial product is able to stimulate autophagy marker ATG16L1, restore number of Paneth cells, and suppress inflammation in colitis.
Figure 7.
Butyrate treatment restores VDR expression in colitis and inhibits inflammation. (A) Butyrate increased VDR protein in IL10−/− mice. IL10−/− mice were given 2% sodium butyrate in the drinking water for 3 weeks. (B) Butyrate increased VDR mRNA level in IL10−/− mice colon. (C) Butyrate increased ATG16L1 mRNA levels in IL10−/− mice. (D) Butyrate decreased IL6 mRNA level. (n=4, * P<0.05).(E) Representative images of indirect immunofluorescence of sections stained for lysozyme (red) in ileal crypts of IL10−/− mice with/without butyrate treatment. Percentage of Paneth cells displayed normal and abnormal (D1 to D3) patterns of lysozyme expression (n=4/group, * P<0.05). (F) A working model of VDR in intestinal and microbiome homeostasis.
Discussion
The positive association of vitamin D deficiency and VDR polymorphisms with IBD suggests that vitamin D metabolism and VDR likely plays a vital regulatory role in this disease.94, 95 Our study and others indicate that VDR deletions exaggerate colitis through activation the NF-κB pathway.25, 26 However the exact role of intestinal VDR in IBD and inflammation remains elusive. In the current study, we established a VDRΔIEC mouse model, with selective knockout of VDR expression in intestinal epithelial cells, to investigate the role of VDR on intestinal inflammation, autophagy, antimicrobial peptide expression, and microbiota composition. This study shows that VDR acts as a master regulator of intestinal homeostasis and establishes a unifying link between VDR, autophagy, the intestinal microbiota, and innate immunity, all factors that have been implicated in the pathogenesis of IBD.
Many studies report a link between autophagy and IBD,96, 97 and in a study of the intracellular bacterium M. tuberculosis, 1,25-dihydroxyvitamin D3 was shown to induce autophagy in human monocytes.61 Our previously published work with Salmonella typhimurium, another intracellular pathogen, indicated that VDR-null mutant mice have worse outcomes with Salmonella-induced infection than wild-type controls.65 Since autophagy is important in host defense and to help limit systemic dissemination of Salmonella, we sought to investigate the specific impact of an intestinal VDR deletion on autophagy. Significantly lower amounts of ATG16L1 were found in VDRΔIEC compared to the VDRloxP/loxP controls at both the protein and transcriptional levels, while leaving the other regulators of autophagy intact (e.g., mTOR, phospho-p70 S6 kinase, Beclin-1, data not shown). ATG16L1 is one of the “core ATG proteins’’, required to form the ATG5-ATG12-ATG16L1 complex to promote elongation and closure of the autophagosome.98 Lack of ATG16L1 led to autophagy deficiency in mouse cells.58, 59 Lysozyme, one of the proteases in lysosomes, is associated with autophagy maturation.99, 100 Our data showed VDR knockout decreased lysozyme in VDR−/− MEF cells and ileal epithelial cells. This data further indicated that loss of VDR protein may result in a defect of autophagy maturation in cells. Others have similarly reported lack of lysozyme staining in the mucus of ATG16L1 mutant mice.13
Lysozyme is also a component and marker of the Paneth cell secretory granule. Paneth cells are specialized epithelial cells primarily located in the small intestine. Paneth cells secrete granules containing antimicrobial peptides including lysozyme.101,102, 103 Our data showed that the total amount and normal expression pattern of Paneth cell decreased in VDR−/− mouse ileum. Defective lysozyme and Paneth cell in VDRΔIEC intestine indicates an abnormality of Paneth cell secretion, particularly in the granule exocytosis pathway. VDR−/− ileum contained an increased proportion of Paneth cells with disorganized or diminished granules or exhibiting diffuse cytoplasmic lysozyme staining. The total amount of Paneth cells also decreased in VDR−/− mice. Interestingly, an ATG16L1 mutation was shown to confer Paneth cell defects in Norovirus-infected mice as well as Crohn’s disease patients.59 Hence, loss of normal ATG16L1 gene function may result in aberrant Paneth cell function, suggesting that Paneth cell dysfunction may play an important role in IBD.59 Another gene linked to IBD, XBP1 has also been implicated in severe Paneth cell and goblet cell dysfunction.104 These studies are consistent with our findings of aberrant Paneth cell function, suggesting that the loss of the ability to secrete antimicrobial peptides may be an important factor in the development of intestinal inflammation and IBD.
The intestinal Paneth cells are known to secrete anti-microbial peptides and shape the composition of the microbiome.87, 88 Paneth cells play a key role in establishing and maintaining the intestinal microbiota, and the ability of Paneth cell alpha defensins to remodel the gut microbiota communities has been well described.105 VDRΔIEC mice were found to have increased bacterial loads and a shift of species (increased E. coli and Bacteroides and decreased butyrate-producing bacteria) and developed a more severe DSS-induced colitis, compared to the VDRloxP/loxP mice, suggesting that microbial dysbiosis in VDRΔIEC mice may sensitize the colonic mucosa to chemical injury induced by DSS. It is possible that a defect in this first line mucosal clearance of microbes may ultimately contribute to excessive chronic inflammatory responses. In the IL10−/− experimental colitis model, we were able to increase intestinal VDR expression by administration of the bacterial metabolite, butyrate, and decrease IL-6 inflammatory cytokine expression, consistent with what has been described by others.106
Our data showed dysbiosis, including decreased abundance of Butyrivibrio, in VDRΔIEC mice increase risk for colitis. Dysbiotic microbial ecology is known to contribute to the pathogenesis of IBD and could be corrected by fecal transplantation, based on a recent study using Nod2-deficient mice69. Our co-housing data indicate that the absence of intestinal epithelial VDR confers a transmissible risk for DSS-induced colitis. Therefore, for the VDR-deficient mice, there are several potential strategies to correct colitis by: 1. treating mice with butyrate, a bacterial natural product; 2. enhancing intestinal VDR expression; and 3. fecal transplantation. We will explore fecal transplantation in a future study.
In skin, cathelicidin is a direct target of VDR and is upregulated by vitamin D.107 Vitamin D is thought to be an important regulator for antimicrobial peptides found in skin, but our study indicates that VDR may be similarly important for AMP expression in the intestine. The regulatory networks that control AMP expression still need to be fully defined, but our findings may have important implications not only for understanding the mechanisms of vitamin D and microbiota remodeling in the gut, but also for other epithelial cell-based autoimmune diseases such as psoriasis 108 where dysbiotic microbial communities and aberrant immune responses may similarly play a role in pathogenesis.
In conclusion, our data indicate that intestinal VDR plays a fundamental role in intestinal homeostasis through its effects on autophagy, on Paneth cells, and on the intestinal microbiota itself. There are mutual interactions between vitamin D, VDR and the microbiome that still remain to be fully understood. However, our study suggests that intestinal VDR may represent a master regulator of inflammation, by regulating autophagy and the production of antimicrobial peptides that, in turn, are responsible for remodeling the bacterial communities that comprise the intestinal microbiota. All of these factors (inflammation, autophagy, microbiota composition) have been implicated in the pathogenesis of IBD; we propose that all of these factors may in fact be regulated under the control of one master regulatory pathway, through VDR (figure 7F). Although the complex regulatory network that controls each of these aspects still needs to be fully elucidated, our findings are important not only for a better understanding IBD but may also have applicability for other autoimmune diseases (such as those of the skin, lung), where the host is in contact with bacteria, and aberrant immune responses and microbiota dysbiosis are implicated. Insights gained from understanding how the VDR pathway is integrally involved in regulating autophagy and changing microbiome diversity may serve as a paradigm for understanding the nature of host defense signals in inflammation.
Significance of this study.
What is already known on this subject?
Vitamin D and the vitamin D receptor (VDR) appear to be important immunological regulators of inflammatory bowel diseases (IBD). Low vitamin D status has been observed in IBD patients. A north-south gradient in rates of Crohn’s disease (CD) suggests that vitamin D deficiency may be an environmental trigger contributing to the pathogenesis of IBD. Moreover, polymorphisms in the VDR gene are associated with susceptibility to IBD. VDR expression is significantly decreased in IBD patients. Bacteria regulate intestinal VDR expression in both gnotobiotic and bacterial-colitis models. Under conditions of intestinal inflammation, VDR negatively regulates bacteria-induced proinflammatory NF-κB activity.
What are the new findings?
Our study for the first time links dysbiosis, innate immune functions (Paneth cells), and genetic susceptibility through intestinal epithelial VDR. Our data provide insights by specifically investigating how intestinal epithelial VDR regulates autophagy and Paneth cells through the autophagy gene ATG16L1, thus changing the microbiome profile. We further show that low levels of VDR correlate with decreased ATG16L1 in the intestine of IBD patients and in an experimental colitis model. Gain- and loss-of- function assays show that VDR transcriptionally regulates ATG16L1, thus decreasing lysozyme. Finally, administration of the bacterial product butyrate increases intestinal VDR and ATG16L1 expression and suppresses inflammation in an experimental colitis model. Our study fills an existing gap by characterizing the precise role of intestinal epithelial VDR in regulating intestinal homeostasis through alterations in intestinal autophagy and the microbiome. It also brings up the possibility that microbial natural products can be use therapeutically to restoration VDR-dependent functions in patients with IBD.
How might it impact on clinical practice in the foreseeable future?
Our studies demonstrate a fundamental relationship between vitamin D, VDR, Paneth cell function, gut microbiota, and genetic susceptibility genes (ATG16L1) that is essential for the maintenance of intestinal homeostasis, but also for the development of chronic states of mucosal inflammation. This knowledge can be immediately used to develop intestinal VDR as a clinical biomarker for identifying patients who might benefit from currently available interventions, as well as for the eventual development of novel strategies for the prevention and treatment of human IBD.
Acknowledgements
We thank Liesbet Lieben for technical assistance with VDRΔIEC mice. This work was supported by the NIDDK (KO1 DK075386 and 1R03DK089010-01), the American Cancer Society (RSG-09-075-01-MBC), and Swim Across America Cancer Research Award to Jun Sun. NIDDK DK42086 (DDRCC), DK097268, and DK47722 to Eugene B Chang.
Footnotes
Conflicts of Interest
The authors have no conflicts of Interest.
References
- 1.Blaney GP, Albert PJ, Proal AD. Vitamin D metabolites as clinical markers in autoimmune and chronic disease. Ann N Y Acad Sci. 2009;1173:384–90. doi: 10.1111/j.1749-6632.2009.04875.x. [DOI] [PubMed] [Google Scholar]
- 2.Adorini L, Penna G. Control of autoimmune diseases by the vitamin D endocrine system. Nat Clin Pract Rheumatol. 2008;4:404–12. doi: 10.1038/ncprheum0855. [DOI] [PubMed] [Google Scholar]
- 3.Grau MV, Baron JA, Sandler RS, et al. Vitamin D, calcium supplementation, and colorectal adenomas: results of a randomized trial. J Natl Cancer Inst. 2003;95:1765–71. doi: 10.1093/jnci/djg110. [DOI] [PubMed] [Google Scholar]
- 4.Gocek E, Studzinski GP. Vitamin D and differentiation in cancer. Crit Rev Clin Lab Sci. 2009;46:190–209. doi: 10.1080/10408360902982128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heaney RP. Vitamin D in health and disease. Clin J Am Soc Nephrol. 2008;3:1535–41. doi: 10.2215/CJN.01160308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannell JJ, Hollis BW, Zasloff M, et al. Diagnosis and treatment of vitamin D deficiency. Expert Opin Pharmacother. 2008;9:107–18. doi: 10.1517/14656566.9.1.107. [DOI] [PubMed] [Google Scholar]
- 7.Campbell FC, Xu H, El-Tanani M, et al. The yin and yang of vitamin D receptor (VDR) signaling in neoplastic progression: operational networks and tissue-specific growth control. Biochem Pharmacol. 2009;79:1–9. doi: 10.1016/j.bcp.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abreu MT, Kantorovich V, Vasiliauskas EA, et al. Measurement of vitamin D levels in inflammatory bowel disease patients reveals a subset of Crohn's disease patients with elevated 1,25-dihydroxyvitamin D and low bone mineral density. Gut. 2004;53:1129–36. doi: 10.1136/gut.2003.036657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim WC, Hanauer SB, Li YC. Mechanisms of disease: vitamin D and inflammatory bowel disease. Nat Clin Pract Gastroenterol Hepatol. 2005;2:308–15. doi: 10.1038/ncpgasthep0215. [DOI] [PubMed] [Google Scholar]
- 10.Sentongo TA, Semaeo EJ, Stettler N, et al. Vitamin D status in children, adolescents, and young adults with Crohn disease. Am J Clin Nutr. 2002;76:1077–81. doi: 10.1093/ajcn/76.5.1077. [DOI] [PubMed] [Google Scholar]
- 11.Haussler MR, Whitfield GK, Haussler CA, et al. The nuclear vitamin D receptor: biological and molecular regulatory properties revealed. J Bone and Mineral Research. 1998;13:325–49. doi: 10.1359/jbmr.1998.13.3.325. [DOI] [PubMed] [Google Scholar]
- 12.Demay MB. Mechanism of vitamin D receptor action. Ann N Y Acad Sci. 2006;1068:204–13. doi: 10.1196/annals.1346.026. [DOI] [PubMed] [Google Scholar]
- 13.Tsoukas CD, Provvedini DM, Manolagas SC. 1,25-dihydroxyvitamin D3: a novel immunoregulatory hormone. Science. 1984;224:1438–40. doi: 10.1126/science.6427926. [DOI] [PubMed] [Google Scholar]
- 14.Adams JS, Hewison M. Unexpected actions of vitamin D: new perspectives on the regulation of innate and adaptive immunity. Nat Clin Pract Endocrinol Metab. 2008;4:80–90. doi: 10.1038/ncpendmet0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aranow C. Vitamin D and the immune system. J Investig Med. 2011;59:881–6. doi: 10.231/JIM.0b013e31821b8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu S, Bruce D, Froicu M, et al. Failure of T cell homing, reduced CD4/CD8alphaalpha intraepithelial lymphocytes, and inflammation in the gut of vitamin D receptor KO mice. Proc Natl Acad Sci U S A. 2008;105:20834–9. doi: 10.1073/pnas.0808700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stio M, Martinesi M, Bruni S, et al. Interaction among vitamin D(3) analogue KH 1060, TNF-alpha, and vitamin D receptor protein in peripheral blood mononuclear cells of inflammatory bowel disease patients. Int Immunopharmacol. 2006;6:1083–92. doi: 10.1016/j.intimp.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 18.Dresner-Pollak R, Ackerman Z, Eliakim R, et al. The BsmI vitamin D receptor gene polymorphism is associated with ulcerative colitis in Jewish Ashkenazi patients. Genet Test. 2004;8:417–20. doi: 10.1089/gte.2004.8.417. [DOI] [PubMed] [Google Scholar]
- 19.Simmons JD, Mullighan C, Welsh KI, et al. Vitamin D receptor gene polymorphism: association with Crohn's disease susceptibility. Gut. 2000;47:211–4. doi: 10.1136/gut.47.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ananthakrishnan AN, Khalili H, Higuchi LM, et al. Higher predicted vitamin D status is associated with reduced risk of Crohn's disease. Gastroenterology. 142:482–9. doi: 10.1053/j.gastro.2011.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes DJ, McManus R, Neary P, et al. Common variation in the vitamin D receptor gene and risk of inflammatory bowel disease in an Irish case-control study. Eur J Gastroenterol Hepatol. 2011;23:807–12. doi: 10.1097/MEG.0b013e328349283e. [DOI] [PubMed] [Google Scholar]
- 22.Eloranta JJ, Wenger C, Mwinyi J, et al. Association of a common vitamin D-binding protein polymorphism with inflammatory bowel disease. Pharmacogenet Genomics. 2011;21:559–64. doi: 10.1097/FPC.0b013e328348f70c. [DOI] [PubMed] [Google Scholar]
- 23.Wada K, Tanaka H, Maeda K, et al. Vitamin D receptor expression is associated with colon cancer in ulcerative colitis. Oncol Rep. 2009;22:1021–5. doi: 10.3892/or_00000530. [DOI] [PubMed] [Google Scholar]
- 24.Froicu M, Zhu Y, Cantorna MT. Vitamin D receptor is required to control gastrointestinal immunity in IL-10 knockout mice. Immunology. 2006;117:310–8. doi: 10.1111/j.1365-2567.2005.02290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu S, Xia Y, Liu X, et al. Vitamin D receptor deletion leads to reduced level of IkappaBalpha protein through protein translation, protein-protein interaction, and post-translational modification. Int J Biochem Cell Biol. 2010;42:329–36. doi: 10.1016/j.biocel.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu W, Chen Y, Golan MA, et al. Intestinal epithelial vitamin D receptor signaling inhibits experimental colitis. J Clin Invest. 2013;123:3983–96. doi: 10.1172/JCI65842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato S. The function of vitamin D receptor in vitamin D action. J Biochem. 2000;127:717–22. doi: 10.1093/oxfordjournals.jbchem.a022662. [DOI] [PubMed] [Google Scholar]
- 28.Gombart AF, Borregaard N, Koeffler HP. Human cathelicidin antimicrobial peptide (CAMP) gene is a direct target of the vitamin D receptor and is strongly up-regulated in myeloid cells by 1,25-dihydroxyvitamin D3. FASEB J. 2005;19:1067–77. doi: 10.1096/fj.04-3284com. [DOI] [PubMed] [Google Scholar]
- 29.Wang TT, Nestel FP, Bourdeau V, et al. Cutting edge: 1,25-dihydroxyvitamin D3 is a direct inducer of antimicrobial peptide gene expression. J Immunol. 2004;173:2909–12. doi: 10.4049/jimmunol.173.5.2909. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Becklund BR, DeLuca HF. Identification of a highly specific and versatile vitamin D receptor antibody. Arch Biochem Biophys. 2009;494:166–77. doi: 10.1016/j.abb.2009.11.029. [DOI] [PubMed] [Google Scholar]
- 31.Bouillon R, Carmeliet G, Verlinden L, et al. Vitamin D and human health: lessons from vitamin D receptor null mice. Endocr Rev. 2008;29:726–76. doi: 10.1210/er.2008-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carlberg C, Seuter S. A genomic perspective on vitamin D signaling. Anticancer Res. 2009;29:3485–93. [PubMed] [Google Scholar]
- 33.Protiva P, Cross HS, Hopkins ME, et al. Chemoprevention of colorectal neoplasia by estrogen: potential role of vitamin D activity. Cancer Prev Res (Phila Pa) 2009;2:43–51. doi: 10.1158/1940-6207.CAPR-08-0103. [DOI] [PubMed] [Google Scholar]
- 34.Murillo G, Mehta RG. Chemoprevention of chemically-induced mammary and colon carcinogenesis by 1alpha-hydroxyvitamin D5. J Steroid Biochem Mol Biol. 2005;97:129–36. doi: 10.1016/j.jsbmb.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 35.Kaler P, Augenlicht L, Klampfer L. Macrophage-derived IL-1beta stimulates Wnt signaling and growth of colon cancer cells: a crosstalk interrupted by vitamin D3. Oncogene. 2009;28:3892–902. doi: 10.1038/onc.2009.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fichera A, Little N, Dougherty U, et al. A vitamin D analogue inhibits colonic carcinogenesis in the AOM/DSS model. J Surg Res. 2007;142:239–45. doi: 10.1016/j.jss.2007.02.038. [DOI] [PubMed] [Google Scholar]
- 37.Nagpal S, Lu J, Boehm MF. Vitamin D analogs: mechanism of action and therapeutic applications. Curr Med Chem. 2001;8:1661–79. doi: 10.2174/0929867013371950. [DOI] [PubMed] [Google Scholar]
- 38.Palmer HG, Sanchez-Carbayo M, Ordonez-Moran P, et al. Genetic signatures of differentiation induced by 1alpha,25-dihydroxyvitamin D3 in human colon cancer cells. Cancer Res. 2003;63:7799–806. [PubMed] [Google Scholar]
- 39.Chan AT, Giovannucci EL. Primary prevention of colorectal cancer. Gastroenterology. 2010;138:2029–43. doi: 10.1053/j.gastro.2010.01.057. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trowbridge R, Mittal SK, Agrawal DK. Vitamin d and the epidemiology of upper gastrointestinal cancers: a critical analysis of the current evidence. Cancer Epidemiol Biomarkers Prev. 2013;22:1007–14. doi: 10.1158/1055-9965.EPI-13-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu PT, Stenger S, Li H, et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science. 2006;311:1770–3. doi: 10.1126/science.1123933. [DOI] [PubMed] [Google Scholar]
- 42.Koslowski MJ, Beisner J, Stange EF, et al. Innate antimicrobial host defense in small intestinal Crohn's disease. Int J Med Microbiol. 2009;300:34–40. doi: 10.1016/j.ijmm.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 43.Wehkamp J, Wang G, Kubler I, et al. The Paneth cell alpha-defensin deficiency of ileal Crohn's disease is linked to Wnt/Tcf-4. J Immunol. 2007;179:3109–18. doi: 10.4049/jimmunol.179.5.3109. [DOI] [PubMed] [Google Scholar]
- 44.Menard S, Forster V, Lotz M, et al. Developmental switch of intestinal antimicrobial peptide expression. J Exp Med. 2008;205:183–93. doi: 10.1084/jem.20071022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lau KS, Cortez-Retamozo V, Philips SR, et al. Multi-scale in vivo systems analysis reveals the influence of immune cells on TNF-alpha-induced apoptosis in the intestinal epithelium. PLoS Biol. 2012;10:e1001393. doi: 10.1371/journal.pbio.1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi N, Vanlaere I, de Rycke R, et al. J Exp Med. 2008;205:1755–61. doi: 10.1084/jem.20080588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adolph TE, Tomczak MF, Niederreiter L, et al. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272–6. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vaishnava S, Behrendt CL, Ismail AS, et al. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A. 2008;105:20858–63. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–35. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–21. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–62. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cadwell K. Crohn's disease susceptibility gene interactions, a NOD to the newcomer ATG16L1. Gastroenterology. 2010;139:1448–50. doi: 10.1053/j.gastro.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 53.Hampe J, Franke A, Rosenstiel P, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–11. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 54.Deretic V, Master S, Singh S. Autophagy gives a nod and a wink to the inflammasome and Paneth cells in Crohn's disease. Dev Cell. 2008;15:641–2. doi: 10.1016/j.devcel.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thachil E, Hugot JP, Arbeille B, et al. Abnormal activation of autophagy-induced crinophagy in Paneth cells from patients with Crohn's disease. Gastroenterology. 2012;142:1097–9. doi: 10.1053/j.gastro.2012.01.031. e4. [DOI] [PubMed] [Google Scholar]
- 56.Kanneganti TD, Lamkanfi M, Nunez G. Intracellular NOD-like receptors in host defense and disease. Immunity. 2007;27:549–59. doi: 10.1016/j.immuni.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 57.Travassos LH, Carneiro LA, Ramjeet M, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 58.Saitoh T, Fujita N, Jang MH, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–8. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 59.Cadwell K, Liu JY, Brown SL, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–63. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verway M, Behr MA, White JH. Vitamin D, NOD2, autophagy and Crohn's disease. Expert Rev Clin Immunol. 2010;6:505–8. doi: 10.1586/eci.10.31. [DOI] [PubMed] [Google Scholar]
- 61.Yuk JM, Shin DM, Lee HM, et al. Vitamin D3 induces autophagy in human monocytes/macrophages via cathelicidin. Cell Host Microbe. 2009;6:231–43. doi: 10.1016/j.chom.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 62.Campbell GR, Spector SA. Vitamin D inhibits human immunodeficiency virus type 1 and Mycobacterium tuberculosis infection in macrophages through the induction of autophagy. PLoS Pathog. 2012;8:e1002689. doi: 10.1371/journal.ppat.1002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun J, Kong J, Duan Y, et al. Increased NF-kappaB activity in fibroblasts lacking the vitamin D receptor. Am J Physiol Endocrinol Metab. 2006;291:E315–22. doi: 10.1152/ajpendo.00590.2005. [DOI] [PubMed] [Google Scholar]
- 64.Sun J, Mustafi R, Cerda S, et al. Lithocholic acid down-regulation of NF-kappaB activity through vitamin D receptor in colonic cancer cells. J Steroid Biochem Mol Biol. 2008;111:37–40. doi: 10.1016/j.jsbmb.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu S, Liao AP, Xia Y, et al. Vitamin D receptor negatively regulates bacterial-stimulated NF-kappaB activity in intestine. Am J Pathol. 2010;177:686–97. doi: 10.2353/ajpath.2010.090998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu S, Sun J. Vitamin D, vitamin D receptor, and macroautophagy in inflammation and infection. Discov Med. 2011;11:325–35. [PMC free article] [PubMed] [Google Scholar]
- 67.Van Cromphaut SJ, Dewerchin M, Hoenderop JG, et al. Duodenal calcium absorption in vitamin D receptor-knockout mice: functional and molecular aspects. Proc Natl Acad Sci U S A. 2001;98:13324–9. doi: 10.1073/pnas.231474698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hamamoto N, Maemura K, Hirata I, et al. Inhibition of dextran sulphate sodium (DSS)-induced colitis in mice by intracolonically administered antibodies against adhesion molecules (endothelial leucocyte adhesion molecule-1 (ELAM-1) or intercellular adhesion molecule-1 (ICAM-1)) Clin Exp Immunol. 1999;117:462–8. doi: 10.1046/j.1365-2249.1999.00985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Couturier-Maillard A, Secher T, Rehman A, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest. 2013;123:700–11. doi: 10.1172/JCI62236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sellon RK, Tonkonogy S, Schultz M, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–31. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu R, Wu S, Liu X, et al. Chronic effects of a Salmonella type III secretion effector protein AvrA in vivo. PLoS One. 2010;5:e10505. doi: 10.1371/journal.pone.0010505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Franks AH, Harmsen HJ, Raangs GC, et al. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1998;64:3336–45. doi: 10.1128/aem.64.9.3336-3345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kochl R, Hu XW, Chan EY, et al. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic. 2006;7:129–45. doi: 10.1111/j.1600-0854.2005.00368.x. [DOI] [PubMed] [Google Scholar]
- 74.Stamatiou R, Paraskeva E, Boukas K, et al. Azithromycin has an antiproliferative and autophagic effect on airway smooth muscle cells. Eur Respir J. 2009;34:721–30. doi: 10.1183/09031936.00089407. [DOI] [PubMed] [Google Scholar]
- 75.Lagishetty V, Misharin AV, Liu NQ, et al. Vitamin D deficiency in mice impairs colonic antibacterial activity and predisposes to colitis. Endocrinology. 2010;151:2423–32. doi: 10.1210/en.2010-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu Z, Shen F, Li X, et al. Molecular and microscopic analysis of bacteria and viruses in exhaled breath collected using a simple impaction and condensing method. PLoS One. 2012;7 doi: 10.1371/journal.pone.0041137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fan Y, Dickman KG, Zong WX. Akt and c-Myc differentially activate cellular metabolic programs and prime cells to bioenergetic inhibition. J Biol Chem. 2010;285:7324–33. doi: 10.1074/jbc.M109.035584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Y, Hoenig JD, Malin KJ, et al. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. The ISME journal. 2009;3:944–54. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Devkota S, Wang Y, Musch MW, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–8. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Masuyama R, Stockmans I, Torrekens S, et al. Vitamin D receptor in chondrocytes promotes osteoclastogenesis and regulates FGF23 production in osteoblasts. J Clin Invest. 2006;116:3150–9. doi: 10.1172/JCI29463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bloom SM, Bijanki VN, Nava GM, et al. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Prindiville TP, Sheikh RA, Cohen SH, et al. Bacteroides fragilis enterotoxin gene sequences in patients with inflammatory bowel disease. Emerging infectious diseases. 2000;6:171–4. doi: 10.3201/eid0602.000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Basset C, Holton J, Bazeos A, et al. Are Helicobacter species and enterotoxigenic Bacteroides fragilis involved in inflammatory bowel disease? Dig Dis Sci. 2004;49:1425–32. doi: 10.1023/b:ddas.0000042241.13489.88. [DOI] [PubMed] [Google Scholar]
- 85.Rabizadeh S, Rhee KJ, Wu S, et al. Enterotoxigenic bacteroides fragilis: a potential instigator of colitis. Inflamm Bowel Dis. 2007;13:1475–83. doi: 10.1002/ibd.20265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kong J, Zhang Z, Musch MW, et al. Novel role of the vitamin D receptor in maintaining the integrity of the intestinal mucosal barrier. Am J Physiol Gastrointest Liver Physiol. 2008;294:G208–16. doi: 10.1152/ajpgi.00398.2007. [DOI] [PubMed] [Google Scholar]
- 87.Kabi A, Nickerson KP, Homer CR, et al. Digesting the genetics of inflammatory bowel disease: insights from studies of autophagy risk genes. Inflamm Bowel Dis. 2012;18:782–92. doi: 10.1002/ibd.21868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–4. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Komatsu M, Waguri S, Koike M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–63. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 90.Zhang XQ, Dunner K, Jr., Benedict WF. Autophagy is induced by adenoviral-mediated interferon alpha treatment in interferon resistant bladder cancer and normal urothelial cells as a cell death protective mechanism but not by the bystander factors produced. Cancer Gene Ther. 2010;17:579–84. doi: 10.1038/cgt.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li XY, Boudjelal M, Xiao JH, et al. 1,25-Dihydroxyvitamin D3 increases nuclear vitamin D3 receptors by blocking ubiquitin/proteasome-mediated degradation in human skin. Mol Endocrinol. 1999;13:1686–94. doi: 10.1210/mend.13.10.0362. [DOI] [PubMed] [Google Scholar]
- 92.Herfarth HH, Mohanty SP, Rath HC, et al. Interleukin 10 suppresses experimental chronic, granulomatous inflammation induced by bacterial cell wall polymers. Gut. 1996;39:836–45. doi: 10.1136/gut.39.6.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Machiels K, Joossens M, Sabino J, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2013 Sep 10; doi: 10.1136/gutjnl-2013-304833. Online First. doi:10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- 94.Mouli VP, Ananthakrishnan AN. Review article: vitamin D and inflammatory bowel diseases. Aliment Pharmacol Ther. 2014;39:125–36. doi: 10.1111/apt.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lu R, Wu S, Xia Y, et al. The Vitamin D Receptor, Inflammatory Bowel Diseases, and Colon Cancer. Current colorectal cancer reports. 2012;8:57–65. doi: 10.1007/s11888-011-0114-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Parkes M. Evidence from genetics for a role of autophagy and innate immunity in IBD pathogenesis. Dig Dis. 2012;30:330–3. doi: 10.1159/000338119. [DOI] [PubMed] [Google Scholar]
- 97.Randall-Demllo S, Chieppa M, Eri R. Intestinal Epithelium and Autophagy: Partners in Gut Homeostasis. Frontiers in immunology. 2013;4:301. doi: 10.3389/fimmu.2013.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hwang S, Maloney NS, Bruinsma MW, et al. Nondegradative role of Atg5-Atg12/ Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe. 2012;11:397–409. doi: 10.1016/j.chom.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Phan TG, Green JA, Gray EE, et al. Immune complex relay by subcapsular sinus macrophages and noncognate B cells drives antibody affinity maturation. Nat Immunol. 2009;10:786–93. doi: 10.1038/ni.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Virgin HW, Levine B. Autophagy genes in immunity. Nat Immunol. 2009;10:461–70. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang D, Peregrina K, Dhima E, et al. Paneth cell marker expression in intestinal villi and colon crypts characterizes dietary induced risk for mouse sporadic intestinal cancer. Proc Natl Acad Sci U S A. 2011;108:10272–7. doi: 10.1073/pnas.1017668108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9:356–68. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 103.Ouellette AJ. Paneth cell alpha-defensins in enteric innate immunity. Cell Mol Life Sci. 2011;68:2215–29. doi: 10.1007/s00018-011-0714-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kaser A, Lee AH, Franke A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–56. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Salzman NH, Hung K, Haribhai D, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schauber J, Svanholm C, Termen S, et al. Expression of the cathelicidin LL-37 is modulated by short chain fatty acids in colonocytes: relevance of signalling pathways. Gut. 2003;52:735–41. doi: 10.1136/gut.52.5.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Segaert S. Vitamin D regulation of cathelicidin in the skin: toward a renaissance of vitamin D in dermatology? J Invest Dermatol. 2008;128:773–5. doi: 10.1038/jid.2008.35. [DOI] [PubMed] [Google Scholar]
- 108.Zeeuwen PL, Kleerebezem M, Timmerman HM, et al. Microbiome and skin diseases. Curr Opin Allergy Clin Immunol. 2013;13:514–20. doi: 10.1097/ACI.0b013e328364ebeb. [DOI] [PubMed] [Google Scholar]