Abstract
Inappropriate activation of the Hh signaling pathway has been implicated in the development of several types of cancers including prostate, lung, pancreas, breast, brain and skin. Present study identified the binding affinities of eight established inhibitors viz., Cyclopamine, Saridegib, Itraconazole, LDE-225, TAK-441, BMS-833923 (XL139), PF-04449913 and Vismodegib targeting SMO receptor - a candidate protein involved in hedgehog pathway and sought to identify the best amongst the established inhibitors through by molecular docking. Exelxis® BMS 833923 (XL 139) demonstrated superior binding affinity aided by MolDock scoring docking algorithm. Further BMS 833923 (XL 139) was evaluated for pharmacophoric features which revealed appreciable ligand receptor interactions.
Keywords: Tumorogenesis, Hedgehog Pathway, SMO Inhibitors, BMS 833923
Background
In the developing embryo, a group of proteins involved in hedgehog pathway send signals that help cells to grow in the right place and in the right way. The hedgehog pathway can also control the growth of blood vessels and nerves. In adults, hedgehog pathway proteins are not usually active, however in cases documented; changes in a gene switch them on. Hedgehog pathway blockers are designed to switch off the proteins and impede the tumor growth. ‘Hedgehog’ proteins (Hh) are secreted signaling proteins that were first discovered in Drosophila, along with many components of their signal transduction machinery [1]. They are highly hydrophobic proteins, which, after secretion, can diffuse and establish gradients in tissues, which have a paramount role in the proper development of the embryo [2].
The first connection between aberrant Hh signalling and cancer was the discovery that the rare condition Gorlin syndrome is caused by a mutation in PTCH1 [3, 4]. Gorlin patients develop numerous basal cell carcinomas (BCCs) during their lifetimes and are predisposed to other kinds of cancer as well, especially medulloblastoma, a tumor of cerebellar granule neuron progenitor cells, and rhabdomyosarcoma, a muscle tumor. More importantly, it was further determined that a large majority of sporadically occurring BCCs also involve hyper activated Hh signalling, as judged by high levels of mRNA of the Hh target genes GLI1 and PTCH1 in tumor cells [5, 6]. Inactivating mutations in PTCH1 occur most commonly in these tumors, with activating mutations in SMOH found in about 10% of all BCCs [7, 8]. These results implicate the Hh pathway as an important pharmacological target for a variety of conditions.
The Hedgehog (Hh) proteins comprise a group of secreted proteins that regulate cell growth, differentiation and survival [9]. They are involved in organogenesis, and promotes adult stem cell proliferation [2, 10]. Inappropriate activation of the Hh signaling pathway has been implicated in the development of several types of cancers including prostate, lung, pancreas, breast, brain and skin [11– 16]. Sonic Hedgehog (Shh) is the best studied ligand of Hh pathway in vertebrates. In the absence of the ligand, the Patched (PTCH) receptor inhibits Smoothened (SMO), a downstream protein in the pathway. Binding of Shh to PTCH alleviates this inhibition, thus regulating the expression of Gli transcription factors [17]. Loss-of function mutations of PTCH, gain-of-function mutations of SMO and misexpression of the Gli2 and Gli3 have been associated with tumor formation and maintenance in animal models of medulloblastoma and basal cell carcinoma of the skin [18–20]. Hedgehog signaling also has an important role in angiogenesis, metastasis and suppression of apoptosis [21–24].
Hedgehog pathway inhibitors are a relatively new class of therapeutic agents that act by targeting the proteins involved in the regulation of Hh pathway. Cyclopamine is the prototype inhibitor of the Shh pathway that inactivates SMO by binding to its hepta-helical bundle [25]. It is currently undergoing preclinical and clinical studies as an anticancer agent in basal cell carcinoma, medulloblastoma and rhabdomyosarcoma [26]. Saridegib (IPI- 926), a synthetic analog of cyclopamine, has shown positive results in phase I clinical trial of advanced solid tumors. Similarly, itraconazole, an antifungal drug, has also been shown to suppress growth of medulloblastoma in mice allograft models [27]. Other candidates for future trials include Novartis׳ LDE-225, Millennium Pharmaceuticals' TAK-441, Exelixis BMS-833923 (XL139) and Pfizer's PF-04449913 [28, 29]. Vismodegib (IPI-926; Erivedge) has been recently approved by the FDA for treatment of advanced basal cell carcinoma [30]. Due to its mechanism of action, it is contraindicated during pregnancy, as it is teratogenic, embryotoxic and fetotoxic (Genentech Inc., 2012).
SMO being candidate protein in hedgehog pathway therefore is a fundamental target in anti tumor drug development. Hence in the view of above, the objective of the present study centers to identify effectual inhibitor amongst the previously stated drugs as aforementioned.
Methodology
Selection of inhibitors:
Inhibitors with their PubChem ID selected for molecular docking is listed in Table 1(see supplementary material).
Preparation of protein and inhibitors:
The structures of selected SMO inhibitors were optimized and cleaned in 3d format using Marvin View (MarvinView 5.6.0.2, 1998-2011, Copyright © ChemAxon Ltd) (Csizmadia, 2000). The three-dimensional structure of SMO [PDB: 4JKV] was retrieved from the Protein Data Bank [31]. The protein was prepared by removing all bound water molecules and ligands. Explicit hydrogens, bond orders, hybridization and charges were assigned to protein structure if missing.
Virtual screening parameters:
Molecular docking program Molegro Virtual Docker (MVD) which includes highly efficient PLP and MolDock scoring function provided a flexible platform for docking all the similar compounds [32– 34]. All the selected SMO inhibitors were docked into inhibitor binding site of SMO in reference to coordinates of bound ligand 1KS_601 (C26 H24 F4N6 O) in the crystal structure of 4JKV as shown in Figure 1. Docking parameters were set to 0.20 Å as grid resolution, maximum iteration of 1500 and maximum population size of 50. Energy minimization and hydrogen bonds were optimized after the docking. Simplex evolution was set at maximum steps of 300 with neighborhood distance factor of 1. Binding affinity of ligand receptor interactions (otherwise depicted by rerank scoring function) was evaluated on the basis of the internal ES (internal electrostatic interaction), internal hydrogen bond interactions and sp2-sp2 torsions.
Figure 1.
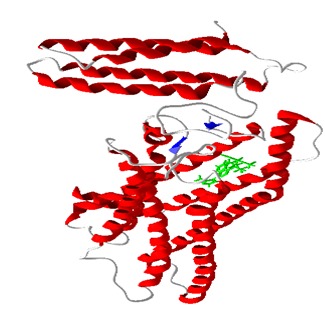
KS_601 (green) bound to SMO in the inhibitory site
Results & Discussion
Interpretation of receptor ligand interactions:
Table 2 (see supplementary material) represents the descending order of docking scores of the inhibitor. In the present investigation BMS 833923 (XL 139) demonstrated superior binding affinity in comparison to remaining inhibitors. Owing to its best binding affinity BMS 833923 (XL 139) was further investigated for its pharmacophoric features.
Structure based pharmacophoric identification of BMS 833923 (XL 139):
Virtual screening of SMO inhibitors identified a molecule BMS 833923 (XL 139) to have best binding affinity against SMO recptor. This candidate showed a better receptor-ligand interaction as evidenced from the MolDock and PLP aided docking scores. Comprehensively shown in Figure 2, the molecule demonstrated van der Waals interactions with Ile389, Phe 484 & 391, Trp 281 Asp 384 & 480, Leu 515, 221 & 522 & 325 Asn 521, Ile 215, 231 & 381, and shows electrostatic interaction with Tyr 394, Gln 477, Arg 400, Glu 518, Asn 219, Tyr 207. Further BMS 833923 (XL 139) showed good hydrogen bond interaction as shown in Figure 3. The hydrogen bonding profile of BMS 833923 (XL 139) is shown in Table 3 (see supplementary material). The overall ligand receptor affinity score i.e. rerank score (a function of stearic interactions, electrostatic interactions, H bond interactions etc.) is shown in Table 4 (see supplementary material). Further BMS 833923 (XL 139) was also evaluated for electrostatic interactions, hydrophobic interactions, and solvent accessible surface area upon ligand binding which is shown in Figure 4 Figure 5& Figure 6respectively.
Figure 2.
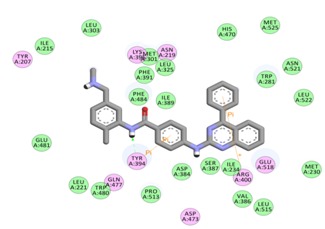
BMS 833923 (XL 139) and its interactions with SMO
Figure 3.
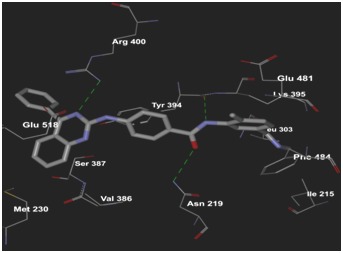
Hydrogen bond interactions of BMS 833923 (XL 139) with SMO
Figure 4.
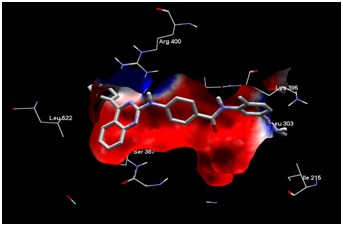
Electrostatic interactions of BMS 833923 (XL 139) with SMO
Figure 5.
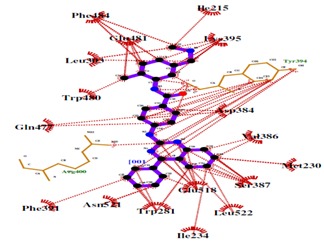
Hydrophobic interactions of BMS 833923 (XL 139) with SMO
Figure 6.
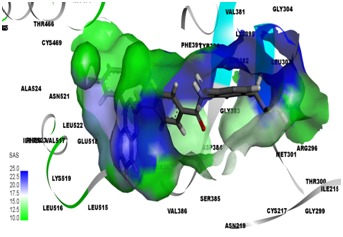
Solvent accessible surface area analysis of BMS 833923 (XL 139) with SMO
Conclusion
Uncontrolled proliferation of cells witnessed in tumor formation is often triggered by the hedge hog pathway that has been once active in embryonic development. Due to environmental and genetic disturbances the pathway is triggered in adult cells leading to tumor formation. Targeting proteins in this pathway has surfaced as an important therapeutic strategy in anticancer drug development. In the present study eight potential inhibitors have been evaluated for its binding affinity against SMO − a G protein coupled receptor which forms a candidate protein in hedgehog pathway. In our study one among the eight established inhibitors, Exelixis׳s ® BMS 833923 (XL 139) demonstrated superior binding affinity with appreciable interactions. Currently this drug is only for research use and undergoing clinical trials. We anticipate from our study that BMS 833923 (XL 139) can be a good nominated drug compared to other existing ones, nevertheless, further research studies are required to support our investigation.
Conflict of interest
The authors confirm that this article content has no conflict of interest.
Supplementary material
Acknowledgments
The financial support from BCIL-DBT (Biotech Industrial Training Programme (BITP) 2013-14) for providing Industry training at Eminent Biosciences, Vijaynagar, Indore, India is gratefully acknowledged.
Footnotes
Citation:Akare et al, Bioinformation 10(12): 737-742 (2014)
References
- 1.Nusslein-Volhard C, et al. Nature. 1980;287:795. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 2.Ingham PW, et al. Genes Dev. 2001;15:3059. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 3.Hahn H, et al. Cell. 1996;85:841. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 4.Johnson RL, et al. Science. 1996;272:1668. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 5.Dahmane N, et al. Nature. 1997;89:876. doi: 10.1038/39918. [DOI] [PubMed] [Google Scholar]
- 6.Unden AB, et al. Cancer Res. 1997;57:2336. [PubMed] [Google Scholar]
- 7.Reifenberger J, et al. Cancer Res. 1998;58:1798. [PubMed] [Google Scholar]
- 8.Xie J, et al. Nature. 1998;391:90. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- 9.Varjosalo M, Taipale J. Genes Dev. 2008;22:2454. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- 10.Bhardwaj G, et al. Nat Immunol. 2001;2:172. doi: 10.1038/84282. [DOI] [PubMed] [Google Scholar]
- 11.Sheng T, et al. Mol Cancer. 2004;3:29. doi: 10.1186/1476-4598-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watkins DN, et al. Nature. 2003;422:313. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 13.Thayer SP, et al. Nature. 2003;425:851. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu S, et al. Cancer Res. 2006;66:6063. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dellovade T, et al. Annu Rev Neurosci. 2006;29:539. doi: 10.1146/annurev.neuro.29.051605.112858. [DOI] [PubMed] [Google Scholar]
- 16.Bale AE, et al. Hum Mol Genet. 2001;10:757. doi: 10.1093/hmg/10.7.757. [DOI] [PubMed] [Google Scholar]
- 17.Michaud EJ, Yoder BK. Cancer Res. 2006;66:6463. doi: 10.1158/0008-5472.CAN-06-0462. [DOI] [PubMed] [Google Scholar]
- 18.Oro AE, et al. Science. 1997;276:817. doi: 10.1126/science.276.5313.817. [DOI] [PubMed] [Google Scholar]
- 19.Nilsson M, et al. Proc Natl Acad Sci U S A. 2000;97:3438. doi: 10.1073/pnas.050467397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grachtchouk M, et al. Nat Genet. 2000;24:216. doi: 10.1038/73417. [DOI] [PubMed] [Google Scholar]
- 21.Lee SW, et al. Int J Mol Med. 2007;19:445. [PubMed] [Google Scholar]
- 22.Li X, et al. Oncogene. 2006;25:609. doi: 10.1038/sj.onc.1209077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adolphe C, et al. Cancer Res. 2006;66:2081. doi: 10.1158/0008-5472.CAN-05-2146. [DOI] [PubMed] [Google Scholar]
- 24.Athar M, et al. Cancer Res. 2004;64:7545. doi: 10.1158/0008-5472.CAN-04-1393. [DOI] [PubMed] [Google Scholar]
- 25.Chen JK, et al. Genes Dev. 2002;16:2743. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taipale J, et al. Nature. 2000;406:1005. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 27.Kim J, et al. Cancer Cell. 2010;17:388. doi: 10.1016/j.ccr.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tremblay MR, et al. Curr Opin Chem Biol. 2010;14:428. doi: 10.1016/j.cbpa.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 29.McMillan R, Matsui W. Clin Cancer Res. 2012;18:4883. doi: 10.1158/1078-0432.CCR-11-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dlugosz A, et al. Nat Rev Drug Discov. 2012;11:437. doi: 10.1038/nrd3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C, et al. Nature. 2013;497:338. doi: 10.1038/nature12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chanda S, et al. Bioinformation. 2014;10:611. [Google Scholar]
- 33.Bandaru S, et al. Bioinformation. 2014;10:652. doi: 10.6026/97320630010652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bandaru S, et al. Curr Top Med Chem. 2013;13:1650. doi: 10.2174/15680266113139990115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


