Abstract
Accurate segregation of duplicated chromosomes between two daughter cells depends on bi-polar spindle formation, a metaphase state in which sister kinetochores are attached to microtubules emanating from opposite spindle poles. To ensure bi-orientation, cells possess surveillance systems that safeguard against microtubule-kinetochore attachment defects, including the spindle assembly checkpoint and the error correction machinery. However, recent developments have identified centrosome dynamics – that is, centrosome disjunction and poleward movement of duplicated centrosomes – as a central target for deregulation of bi-orientation in cancer cells. Here we review novel insights into the mechanisms that underlie centrosome dynamics and discuss how these mechanisms are perturbed in cancer cells to drive chromosome missegregation and advance neoplastic transformation.
Keywords: Centrosome disjunction, centrosome movement, centrosome dynamics, chromosomal instability, cancer
A CLINICAL PERSECTIVE ON CENTROSOME DYNAMICS
Whole chromosome instability (W-CIN), the inability to faithfully segregate duplicated chromosomes between two daughter cells, can result in cells with abnormal numbers of chromosomes, a condition referred to as aneuploidy [1]. Although a causal relationship between aneuploidy and cancer remains a topic of debate, aneuploidy is a common feature of human cancers and one of several mechanisms by which aneuploidy is thought to drive tumorigenesis is through loss of whole chromosomes that contain key tumor suppressor genes [1, 2]. Nonetheless, the molecular and genetic bases of W-CIN in human cancer cells remain poorly understood. While much attention has been focused on the role of mitotic checkpoint signaling, attachment error correction, and centrosome duplication, mutations in key regulators of these mitotic processes are rarely observed in human cancer [3]. Recently, improper timing of centrosome separation has been appreciated as a new and potentially frequent source of aneuploidization in human cancers [4–7]. In this review, we highlight novel insights into normal centrosome biology and explain how abnormal centrosome dynamics are thought to drive chromosome segregation errors and promote cancer.
CENTROSOME STRUCTURE
In animal cells, the centrosome is the primary microtubule-organizing center [8, 9]. In non-dividing cells, centrosomes are located in close proximity to the nucleus where they organize microtubules to help establish cell shape, polarity and proper positioning of subcellular organelles [10]. In dividing cells, centrosomes duplicate to form a bipolar mitotic spindle that separates chromosomal content evenly between two daughter cells. [9]. One centrosome consists of two orthogonally positioned cylindrical organelles, called centrioles that are joined by fibers connecting their proximal ends. Centrioles are surrounded by an unstructured mass of proteins referred to as the pericentriolar material (PCM) [11]. Each centriole consists of nine sets of microtubule triplets assembled into a cartwheel structure. Paired centrioles differ from each other in that one is the mother centriole with appendages at its distal ends and the other is the daughter centriole, which lacks these structures. The PCM contains γ-tubulin ring complexes (γ-TuRCs) as well as several large coiled-coil proteins including pericentrin, ninein and Cep135, which together function in nucleating, anchoring, and positioning microtubules [12].
THE CENTROSOME CYCLE
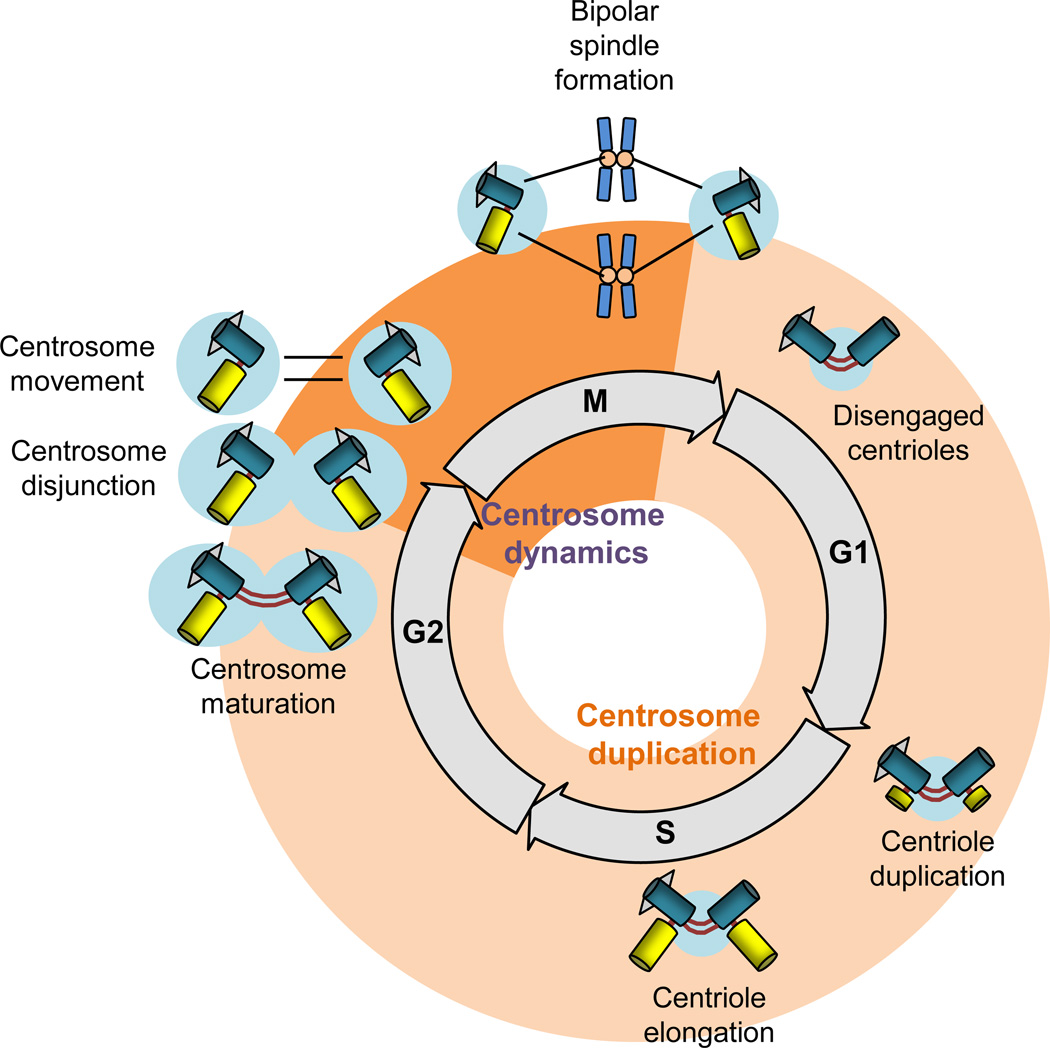
The centrosome is duplicated and separated once per cell cycle and proceeds in a timely fashion that is coordinated with the cell cycle. For the purposes of this review, we will divide the centrosome cycle into two parts: centrosome duplication and centrosome dynamics, which encompasses centrosome splitting (disjunction) and movement. Centrosome duplication takes place between late G1 and early G2 phase and is characterized by four stages: centriole disengagement, centriole duplication, centriole elongation, and centrosome maturation. The key events and molecular drivers of each of the four stages are summarized in Box 1. Additional background on centrosome duplication can be found in several recent in-depth reviews [13–16]. Part two of the centrosome cycle, centrosome dynamics, takes place between late G2 and early M phase and will be reviewed in detail below.
BOX 1. The four stages of centrosome duplication.
Centriole disengagement
During late mitosis and early G1 phase, the perpendicular orientation of centriole pairs is lost as the daughter centriole begins to dissociate and remains attached to the mother centriole merely through flexible fibers (Figure I). This process, termed centriole disengagement, is essential for limiting centriole duplication to one round per cell cycle and for recruiting PCM [17]. Separase and polo-like kinase 1 (Plk1) have been identified as key regulators of centriole disengagement [18]. Separase is thought to drive disengagement by removing cohesin rings that hold mother and daughter centrioles together through proteolytic cleavage of Scc1 [19]. However, subsequent publications have argued against a need for cleavage of centriole-associated cohesin in disengagement [20, 21]. Plk1 regulates centriole disengagement through its interaction with a centrosomal splice variant of Sgo1 known as shorter shugoshin 1 (sSgo1). sSgo1 protects centriolar cohesin from untimely separase-mediated cleavage through a direct interaction with Plk1 [22]. Astrin and Akt kinase interacting protein 1 (Aki1 kinase) also regulate centromeric and centriolar cohesin dissolution through a largely unresolved mechanism [23, 24].
Centriole duplication
Following disengagement, the cell contains one centrosome comprised of two structurally and functionally dissimilar centrioles. The mother centriole, which originates from the mother cell, nucleates and organizes microtubules, whereas the daughter centriole, which is replicated from the mother centriole as a procentriole, only nucleates microtubules and is relatively mobile [25]. Synthesis of the daughter centriole from a pre-existing mother centriole, termed centriole duplication, takes place in early S phase. Centriole duplication is incompletely understood, but involves at least five key proteins: CEP192, SAS-4 (CPAP), SAS-5 (STIL), SAS-6 and Plk4 [26]. The latter two are particularly important as their overexpression is associated with increased centriole synthesis, centrosome amplification, and chromosomal instability [27–35]
Centriole elongation
Newly generated daughter centrioles (also called procentrioles) that emerge near the proximal ends of mature centrioles during early S phase elongate and reach full length by late G2 or early M phase (Figure I). Elongation requires several proteins, including POC5, OFD1, SAS-4, and CP110 [14], with the latter two being best understood at a molecular level [36–38].
Centrosome maturation
In late G2, the PCM dramatically increases γ-TuRCs and its associated proteins, a process referred to as centrosome maturation (Figure I) [39]. Both Plk1 and Aurora-A are recruited to the centrosomes in G2 to mediate maturation [40, 41]. Recruitment of Aurora-A to centrosomes is dependent on Plk1 activity, while activation of centrosome-associated Plk1 is dependent on Aurora-A kinase activity [42]. The microtubule-binding protein TPX2 is also involved in centrosome maturation, presumably by interacting with, and activating Aurora-A [43–45]. Other Plk1 substrates implicated in maturation are pericentrin, NEDD1, CEP192, and CEP215 [46]. Centrosome maturation is initiated when Plk1 phosphorylates pericentrin and NEDD1, which properly targets γ-TuRC to the PCM [47, 48]. Aurora-A then localizes to the centrosome and is activated by Plk1 and the LIM kinase protein Ajuba to complete centrosome maturation by recruiting LATS2, NDEL1 and TACC3 to the centrosome via phosphorylation [42].
Text Box 1; Figure I. The centrosome cycle. The centrosome cycle is divided into two parts: centrosome duplication, which encompasses centriole disengagement, duplication, elongation, and maturation, and centrosome dynamics, which includes centrosome disjunction and movement. In late mitosis, the daughter centriole disengages from the mother centriole and the two centrioles lose their orthogonal configuration (centriole disengagement). At the G1/S transition, one new daughter centriole is synthesized per mother centriole to form a new centrosome (centriole duplication and elongation). In late G2, both centrosomes recruit PCM components in order to prepare for mitosis (centrosome maturation). At the G2/M transition, centrosomal cohesion is gradually lost (centrosome disjunction) and centrosomes move toward opposite poles to form a bipolar spindle (centrosome movement) [9–11].
CENTROSOME DYNAMICS
Centrosome dynamics can be subdivided into two stages: centrosome disjunction in late G2, and microtubule motor protein-mediated centrosome movement toward opposite poles in prophase or prometaphase.
Centrosome disjunction
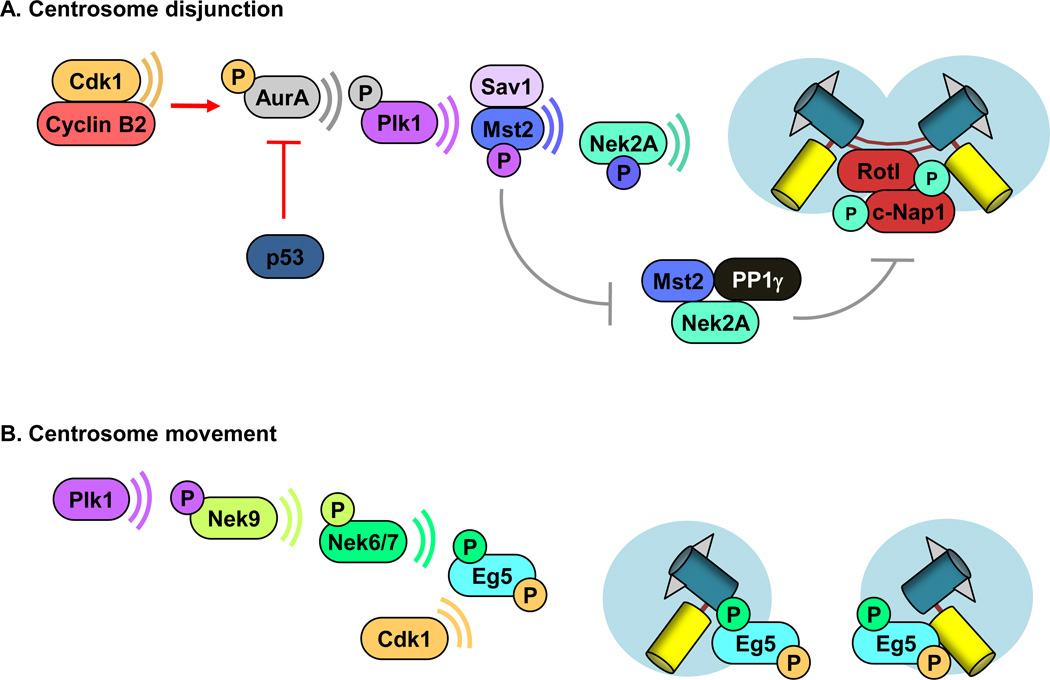
Until late G2, centrosomes are joined by centrosomal cohesin complexes, which include C-Nap1 and rootletin [49]. C-Nap1 is a large coiled-coil protein that localizes rootletin to the proximal end of centrioles, which physically links the two centrioles via fibrous polymers [49]. Late in G2, C-Nap1 and rootletin are phosphorylated by the NIMA-related S/T kinase Nek2A, causing centrosome disjunction (Figure 1A) [50, 51]. Accumulation of Nek2A to centrosomes is regulated by two components of the Hippo pathway, hSav1 and Mst2, the latter of which activates Nek2A through phosphorylation (Figure 1A) [52]. In turn, Plk1 activates Mst2 through phosphorylation. This particular post-translational modification also promotes centrosome disjunction by preventing Nek2A binding to PP1γ a phosphatase, which antagonizes Nek2A (Figure 1A).
Figure 1. Centrosome dynamics.
Centrosome separation occurs in two parts from late G2 through mitosis: disengagement and movement. (A) Hypothetical model for centrosome disjunction. Centrosome-associated cyclin B2/Cdk1 acts to initiate centrosome disjunction through phosphorylation of Aurora A, which in turn phosphorylates Plk1. Activated Plk1, phosphorylates Sav1-bound Mst2, triggering Mst2-mediated phosphorylation of Nek2A. Activated Nek2A phosphorylates the centrosome linker proteins C-Nap1 and rootletin, causing the physical separation of sister centrosomes. PPlγ antagonizes Nek2A phosphorylation of C-Nap1 and Rootletin. [13, 52, 57]. (B) Model for centrosome movement. Plk1 targets Eg5 to centrosomes through sequential phosphorylation of Nek9 and Nek6/7. Cdk1 has also been proposed to be important for Eg5 activation and binding to microtubules [13, 57, 63, 93].
Recently, it was discovered that cyclin B2/Cdk1 is an upstream regulator of the Nek2-dependent centrosome disjunction pathway [5]. Cyclin B2 is a member of the B-type cyclin family and activates Cdk1 through direct binding [53]. Prior to this study, little was known about cyclin B2 other than it localized to centriolar satellites in somatic cells and had an undefined role in spindle formation in Xenopus oocytes [54, 55]. In experiments using gain- and loss-of-function models, cyclin B2 was found to play a critical role in centrosome separation during G2 by promoting Aurora A activity at centrosomes (Figure 1A). Aurora-A is required for the activation of Plk1 [56], which, as mentioned, regulates centrosome separation by Nek2A-dependent phosphorylation of the centrosome linker proteins C-Nap1 and rootletin [13, 57]. Overexpression of cyclin B2 triggers hyperactivation of this pathway, resulting in premature centrosome separation. Conversely, depletion of cyclin B2 reduced centrosomal p-Aurora-A and p-Plk1 and inhibited centrosome disjunction in mouse and human primary cells. Control of centrosome separation by cyclin B2 is dependent on p53, which negatively regulates Aurora-A both transcriptionally and post-transcriptionally [58]. Loss of p53 increases Aurora-A expression and phosphorylation, resulting in premature centrosome separation. This finding indicates that cyclin B2 and p53 antagonistically coordinate centrosome disjunction and demonstrate that cyclin B2/Cdk1 is the upstream regulator of the Nek2A-dependent centrosome disjunction pathway.
The Wnt signaling effector β-catenin has also been implicated in regulating centrosome disjunction [59]. β-catenin was found to be a substrate of Nek2A and localize to proximal and distal regions of centrioles [60]. Depletion of β-catenins lead to monopolar spindles, underscoring its role in centrosome dynamics [59]. Depletion of either rootletin or C-Nap1 causes precocious centrosome disjunction, indicating that β-catenin operates in a functionally distinct manner.
In addition to C-Nap1 and rootletin, Cep68 and Cep215 were identified as centrosomal linker proteins in an siRNA-based screening approach for centrosome cohesion [61]. Both Cep68 and Cep215 are found at centrosomes, but differ in their precise localization. While Cep68 contributes to a dynamic linker structure, Cep215 interacts with and is thought to function downstream of pericentrin [61]. The loss of Cep68 from centrosomes upon mitotic entry of cells and its reliance upon the presence of other linker proteins for recruitment to centrosomes is consistent with Cep68 being a target of Nek2A. The differing behavior and localization of Cep215 favors an alternative mode of regulation, perhaps directly through Plk1.
Centrosome movement
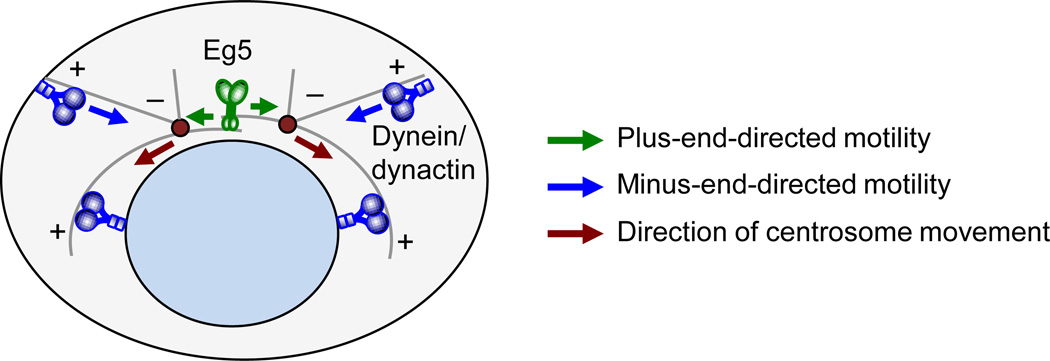
After centrosome disjunction (at G2/M transition), the two untethered centrosomes begin to move toward their respective anchoring sites. Centrosome movement is a complex, multifaceted process that remains incompletely understood at the molecular level [13, 62]. Eg5 plays a central role in centrosome movement. This plus-end directed microtubule motor protein accumulates at centrosomes and astral microtubules, where it generates an outward “pushing” force by sliding antiparallel microtubules between centrosome pairs in opposite directions (Figure 2). Nek6 and 7, and presumably cyclin-Cdk1, facilitate Eg5 targeting to centrosomes through phosphorylation (Figure 1B) [63]. Activation of Nek6 and 7 is mediated by Nek9, which in turn is activated by Plk1 kinase activity [63–66]. While the importance of Eg5 in poleward movement of centrosomes has been firmly established, recent studies have demonstrated that additional proteins are also required in order to generate the necessary forces for centrosome movement [62, 67]. Dynein/dynactin motor complexes at the cell cortex and the nuclear envelope generate a “pulling” force to properly position individual centrosomes (Figure 2) [68, 69]. Additionally, kinetochores are implicated in centrosome movement through the formation of K-fibers [70]. Moreover, the actin cytoskeleton was found to be involved in centrosome movement [62]. However, how actin generates the necessary forces for centrosome movement remains largely unclear.
Figure 2. Motor proteins forces drive poleward movement of duplicated centrosomes.
Separated centrosomes move toward their respective anchoring sites with the help of multiple motor proteins, including Eg5 and dynein. Eg5 is a plus-end-directed motor protein that generates an outward pushing force between the centrosome pair [62]. Cortical and nuclear envelope associated dynein pull centrosomes apart through minus-end-directed force [94, 95].
ABNORMAL CENTROSOME DYNAMICS
The relationship between centrosome separation and chromosome missegregation has been historically overlooked for several reasons. First, bipolar spindle formation can be achieved in some experimental settings when centrosomes are removed via laser ablation or microsurgery [71, 72], leading some to question their role in the process. Second, initiation of centrosome separation within individual cells of a culture has been reported to occur over a range of time spanning prophase to prometaphase, implying flexibility between the synchronization of centrosome separation and mitotic progression [70, 73]. Furthermore, centrosomes that are immobilized prior to NEBD can compensate for this delay by separating their centrosomes in prometaphase without overtly compromising chromosome segregation [74, 75]. Third, Drosophila melanogaster lacking centrosomes can develop without any conspicuous abnormalities [76]. Therefore, irregularities in the control of centrosome splitting and movement prior to NEBD have been largely neglected as a potential source of aneuploidization. However, following several recent advances, it is becoming increasingly apparent that centrosome separation is a highly orchestrated process that needs to unfold in a timely and controlled fashion in order to create a bipolar spindle that accurately segregates duplicated chromosomes between two daughter cells.
Abnormal centrosome dynamics and aneuploidization
As aforementioned, recent work from several laboratories now points to a critical role of proper centrosome dynamics in ensuring accurate segregation of sister chromatids, with both delayed and accelerated centrosome separation increasing rates of spindle geometry defects in metaphase and mitotic errors [4, 5, 7, 77] (Figure 3). Furthermore, evidence is mounting that centrosome dynamics are tightly regulated to limit the risk of neoplastic transformation, which will be discussed in greater detail in the next section. However, we will first review the recent advances made in understanding how delayed centrosome separation leads to W-CIN and discuss potential models for how accelerated centrosome separation might also promote mitotic errors.
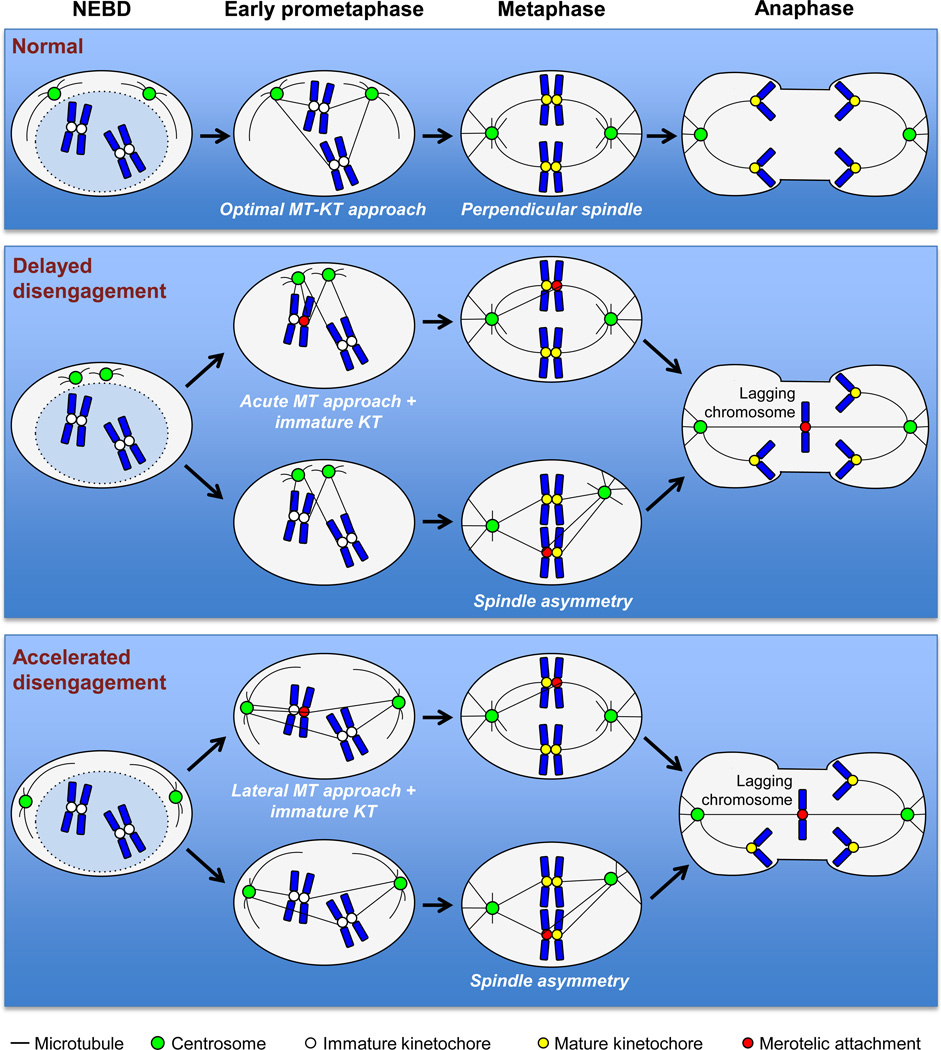
Figure 3. Hypothetical mechanisms by which aberrant centrosome dynamics promote merotely and chromosome lagging.
Normal centrosome dynamics promote amphitelic attachments and faithful chromosome segregation between daughter cells. Delayed centrosome disengagement promotes merotelic attachments in early prometaphase because kinetochores are accessible to microtubules from spindle poles in close proximity. At this stage, immature kinetochores have not yet recruited all of the proteins necessary for proper kinetochore-microtubule attachments, increasing the likelihood of merotelic attachments and/or decreasing the efficiency at which merotelic attachments are detected and resolved. Alternatively, delayed centrosome disengagement is associated with spindle asymmetry in metaphase, which may also promote misattachments prior to anaphase. Accelerated centrosome disengagement may promote merotelic attachments in early prometaphase again through a combination of suboptimal microtubule approach angle to the kinetochore and immature kinetochores that cannot detect and/or resolve merotelic attachments. Like delayed disengagement, accelerated centrosome disengagement has been associated with spindle asymmetry in metaphase, which is likely to promote merotelic attachments just prior to anaphase onset. Unresolved merotelic attachments resolve in chromosome lagging. MT; microtubule. KT; kinetochore.
Pioneering work in understanding how delayed centrosome separation facilitates chromosome missegregation has come from the work of Silkworth and Cimini [77]. Using a subclone of Ptk1 cells with slow separating centrosomes, they uncovered that microtubules emanating from spindle poles in close proximity to each other promote merotelic attachments, a type of attachment error in which one kinetochore is attached to microtubules originating from both spindle poles (Figure 3). These merotelic attachments can form either directly or indirectly through a syntelic intermediate, where both sister kinetochores are attached to microtubules from a single centrosome. Merotelic attachments are particularly dangerous for dividing cells because they are not recognized by the spindle assembly checkpoint and can result in chromosome lagging if left uncorrected by an overwhelmed error correction machinery (Figure 3) [78]. This principle was later illustrated in Usp44 knockout mouse embryonic fibroblasts (MEFs), where delayed centrosome splitting led to abnormal spindle positioning in metaphase and lagging chromosomes [4].
Recent studies in cyclin B2 overexpressing cells have revealed that accelerated centrosome separation can also culminate in abnormal spindle positioning in metaphase, the formation of merotelic attachments, and lagging chromosomes [5]. While this study indicates that centrosome movement and microtubule-kinetochore attachment need to be tightly coordinated to ensure faithful chromosome segregation, the mechanistic basis for increased merotely upon accelerated centrosome splitting remains currently unknown.
One possibility is that early in prometaphase, the immature kinetochores of cells with completely (or almost completely) separated centrosomes are less susceptible to capturing laterally-approaching microtubules and are therefore prone to forming merotelic attachments (Figure 3). After NEBD, kinetochore assembly is still ongoing and cytoplasmic kinetochore proteins that are important for proper microtubule-kinetochore attachment, like BubR1, are being actively recruited to kinetochores. Mature kinetochores are massive, multi-protein structures that may promote proper microtubule attachments and prevent improper attachments in part due to steric hindrance. When a microtubule emanating from a spindle pole is laterally approaching an immature kinetochore, the immature kinetochore may be insufficiently developed to direct the microtubule to its optimal position at the unformed outer kinetochore and, therefore, the microtubule may be just as likely to bind improperly to the opposite kinetochore and form a merotelic attachment.
Another possibility may be that elevated rates of merotely are a consequence of the spindle geometry defects resulting from early centrosome disengagement (Figure 3). Perpendicular spindle geometry is defined as a roughly 90° angle between the metaphase plate and the imaginary line connecting the two spindle poles positioned on opposite sides of the metaphase plate (Figure 4). Spindle poles that are not positioned perpendicular to the metaphase plate may promote merotelic attachments that could effectively overwhelm the error correction machinery and reduce its ability to correct merotelic attachments prior to anaphase.
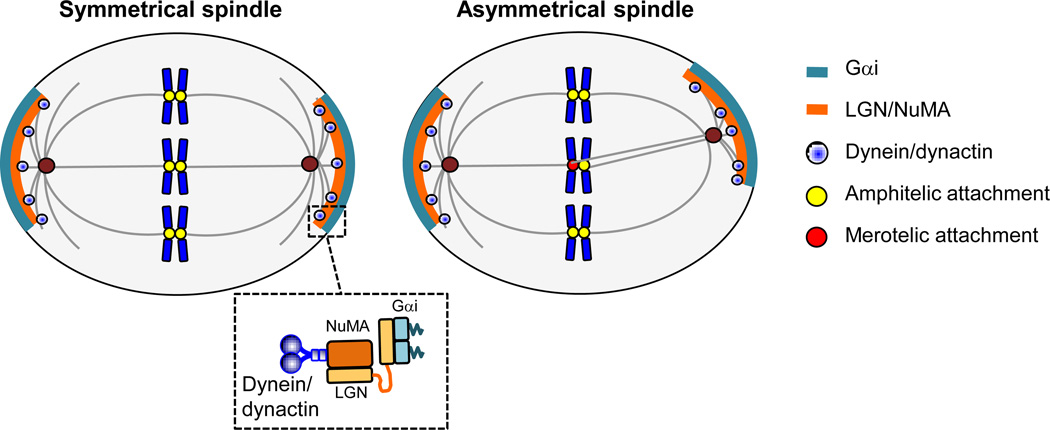
Figure 4. Improper anchoring of astral microtubules at cell cortex as a source of spindle geometry defects.
In cells with normal spindle geometry (left), dynein/dynactin interacts with the Gαi-LGN-NuMA anchor protein complex [89] and captures astral microtubules near the cortex contributing to symmetrical spindle pole orientation and proper kinetochore-microtubule attachment. In cells with abnormal spindle geometry, demarcation of cortical anchoring regions is not symmetrical, resulting in improper kinetochore-microtubule attachment (right).
Alternately, the Aurora B-driven error correction machinery may function less efficiently in correcting merotely in the presence of aberrant spindle forces generated at inner centromeres of asymmetric spindles (Figure 3). Inner centromeric Aurora B destabilizes faulty kinetochore-microtubule attachments, including merotelic attachments, by phosphorylating its substrates located on outer kinetochores. Recent studies suggest that Aurora B kinase activity is dependent on its proximity to these substrates [79–81]. Because intrakinetochore stretch appears to function as an error correction switch [82, 83] – low stretch in the absence of tension will promote destabilization whereas high stretch in the presence of tension will promote stable attachments – the tension generated under asymmetric spindle conditions may inappropriately silence Aurora B kinase activity by physically dissociating it from its substrates, leaving merotelically attached kinetochores unresolved.
As mentioned previously, centrosome dynamics encompass both disengagement and poleward movement. It is conceivable that altered centrosome movement, either too slow or too rapid, also acts to promote merotelic attachment and chromosome lagging. The molecular mechanisms driving centrosome movement remain poorly understood, which is a key barrier to the thorough testing of this idea. Currently, Eg5 is perhaps the most obvious candidate for a molecular target that would slow centrosome movement and impair establishment of an orthogonal bipolar spindle. In addition to Eg5-specific mutations that might limit its motor potential or impair its microtubule binding ability, defective centrosomal targeting of Eg5 might also delay poleward movement and result in abnormal spindle geometry. Similarly, overexpression of Eg5 may also negatively impact centrosome dynamics and promote chromosome segregation errors. In line with this idea, MEFs that overexpress Eg5 were found to be genomically unstable and were unable to form proper bipolar spindles in mitosis [84]. Furthermore, Eg5 transgenic mice were highly prone to spontaneous tumor formation, further linking centrosome separation defects, W-CIN, and cancer.
Intriguingly, both delayed and accelerated centrosome separation promote formation of non-symmetrical mitotic spindles with improperly oriented poles [4, 5]. One possible explanation could be that improper timing of centrosome movements causes deregulation of molecular events that mediate spindle pole anchoring at the cell cortex (Figure 4). Much of the current mechanistic knowledge about cortical anchoring of spindle poles comes from stem cells, which exploit anchoring to specific cortical regions to mediate unequal separation of proteins and organelles during cell division, allowing one daughter cell to retain self-renewal capacity and the other to engage in cell differentiation [85–87]. Cortical anchoring sites are created by specific multi-protein complexes. The G protein Gαi accumulates at designated anchoring sites and mobilizes LGN, a large scaffold protein. LGN in turn is responsible for recruitment of the microtubule binding protein NuMA and the motor complex dynein/dynactin, which provides plus end directed microtubule-pulling forces (Figure 4) [85, 88]. Core components of the spindle pole anchoring machinery are also present in cells, where they may play a central role in spindle orientation and anchoring as well [88, 89]. It will therefore be interesting to explore whether these components are subject to deregulation in cells with spindle geometry defects due to abnormal spindle pole anchoring. Furthermore, signals from the extracellular matrix are known to contribute to spindle orientation, for instance by regulating cortical actin structure and dynamics [90–92], and should also be further interrogated with regards to spindle geometry defects associated with untimely centrosome separation.
Abnormal centrosome dynamics and cancer
The physiological relevance of incomplete centrosome separation was recently demonstrated in mice lacking the Cdc20 deubiquitinase Ups44 [4]. Usp44 knockout mice were prone to aneuploidization and highly susceptible to spontaneous tumor formation. Usp44 loss caused chromosome lagging and Usp44 was subsequently found to be required for proper centrosome disjunction and perpendicular spindle formation in metaphase (Figure 5). Conversely, accelerated centrosome separation in G2 phase seems to be a key tumor-promoting event in mice that overexpress cyclin B2 [5]. High levels of cyclin B2 were found to promote centrosomal aurora A and Plk1 hyperactivity, which resulted in abnormal spindle geometry and W-CIN characterized predominantly by lagging chromosomes (Figure 5). Moreover, knockdown of cyclin B2 in non-transformed primary cells led to reduced activity of aurora A and Plk1, incomplete centrosome separation, asymmetric metaphase spindle geometry, and chromosome lagging.
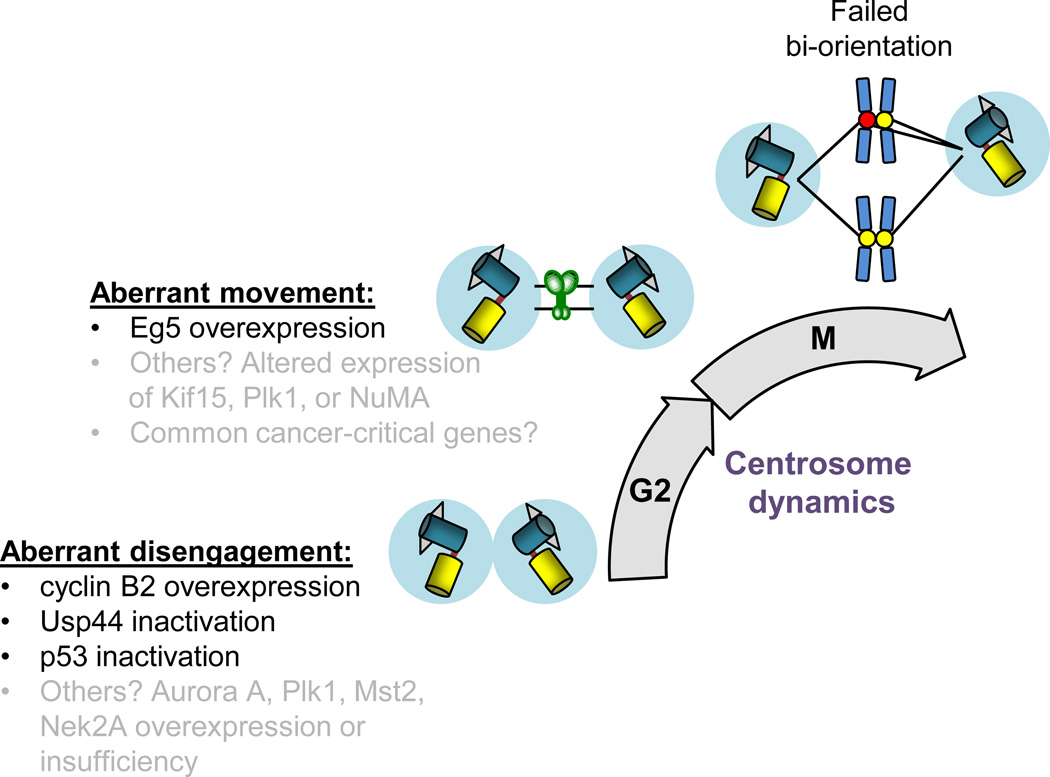
Figure 5. Cancer-associated molecular drivers of abnormal centrosome dynamics.
Usp44 knockout and cyclin B2 overexpression cause spontaneous tumors in mice and feature delayed and accelerated centrosome separation, respectively [4, 5]. p53 inactivation also causes abnormal centrosome disjunction and is the most commonly mutated tumor suppressor in human cancers. It is likely that there are many other proteins, such as the CIN70 gene Nek2A [96], that regulate centrosome disjunction that, when altered, promote CIN and may be associated with cancer. Likewise, abnormal centrosome movement, as is the case for Eg5 overexpression, is tightly associated with CIN and tumorigenesis. There remains a critical need to identify other cancer-associated genes that normally regulate centrosome dynamics.
Taken together, these studies imply a critical role for proper timing of centrosome separation in tumor suppression. Both incomplete and accelerated centrosome separation are tightly associated with tumorigenesis in vivo, raising the question of how common abnormal centrosome separation is in human cancers without centrosome amplification. Because centrosome dynamics are so sensitive to perturbations – that is, both premature and delayed separation result in improperly positioned spindles and W-CIN – one can envision centrosome separation defects being a common hallmark of genetically unstable cancer cells. The observation that lagging chromosomes represent the main segregation defect in cancerous cells [78] supports this notion. Indeed, p53 was discovered to govern proper timing of centrosome separation and three-dimensional spindle positioning by regulating centrosomal aurora A activity [5, 58]. The implication of this discovery is that tumors characterized by p53 loss-of-function may adopt a mutator phenotype in part due to whole chromosome loss or gain resulting from early centrosome disjunction and formation of asymmetric mitotic spindles (Figure 5). In this way, p53 is a prototypical example of a commonly altered cancer-critical gene that can also drive CIN. In fact, several tumor suppressors and oncogenes have been reported to induce CIN [3]. It will be important to determine which kinds of p53 mutations found in human cancers perturb aurora A control in order to better understand the prevalence of abnormal centrosome dynamics in human cancers. Likewise, it will be important to discern which other commonly altered tumor suppressor proteins and/or oncogenes drive CIN by altering normal centrosome dynamics.
CONCLUDING REMARKS
The centrosome organizes microtubules within the cell and helps establish cell shape, polarity, subcellular localization of organelles, and bipolar spindle formation. The centrosome is duplicated, disjoined, and translocated once per cell cycle in distinct phases that are governed by a subset of master regulators that include Plk1, Plk4/SAS-6, aurora A, Eg5/dynein, cyclin B2, and p53. Recent advances in the field have implicated normal centrosome dynamics as a critical process to ensure accurate segregation of duplicated chromosomes between daughter cells. Furthermore, studies aimed to decipher the physiological relevance of centrosome dynamics in mice have suggested that abnormal timing of centrosome separation may be a common and potent CIN-inducing event tightly associated with cancer. These studies have shown that proper centrosome splitting may be a commonly deregulated process in human cancers by virtue of the fact that the most frequently mutated tumor suppressor, p53, plays a central role in governing centrosome disjunction by controlling aurora A activity. This observation raises the possibility that other frequently mutated cancer-critical tumor suppressor genes and oncogenes may promote W-CIN by disrupting the timing and/or movement of centrosomes as the cell attempts to form a bipolar spindle. The frequency of centrosome separation defects in cancer cells and the molecular pathways implicated therein will be important to identify in order to better understand the stepwise process of cellular transformation, pathogenesis of cancer, and to develop novel therapeutic approaches for patients with malignant neoplasms.
Highlights.
The relationship between centrosome separation and chromosome missegregation has historically been overlooked
Gene mutations that delay or accelerate centrosome separation cause spindle malformation, merotely, aneuploidy and cancer
Cyclin B2 and p53 play a central role in centrosome separation
Centrosome separation defects may be a frequent source of chromosomal instability in human cancers
Acknowledgements
We thank Paul Galardy, Darren Baker, Janine van Ree and Arun Kanakkanthara for critical reading of the manuscript and providing helpful comments and discussion. R.M.N. (F30 CA189339-01 and T32 GM 65841) and J.M.v.D. (CA126828) were supported by grants from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Ricke RM, van Deursen JM. Aneuploidy in health, disease, and aging. The Journal of Cell Biology. 2013;201:11–21. doi: 10.1083/jcb.201301061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker DJ, et al. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16:475–486. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orr B, Compton DA. A double-edged sword: how oncogenes and tumor suppressor genes can contribute to chromosomal instability. Frontiers in oncology. 2013;3:164. doi: 10.3389/fonc.2013.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, et al. USP44 regulates centrosome positioning to prevent aneuploidy and suppress tumorigenesis. The Journal of clinical investigation. 2012;122:4362–4374. doi: 10.1172/JCI63084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nam HJ, van Deursen JM. Cyclin B2 and p53 control proper timing of centrosome separation. Nat Cell Biol. 2014;16:538–549. doi: 10.1038/ncb2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaseda K, et al. Dual pathway spindle assembly increases both the speed and the fidelity of mitosis. Biology open. 2012;1:12–18. doi: 10.1242/bio.2011012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silkworth WT, et al. Timing of centrosome separation is important for accurate chromosome segregation. Mol Biol Cell. 2012;23:401–411. doi: 10.1091/mbc.E11-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 9.Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nature reviews. Molecular cell biology. 2007;8:451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- 10.Nigg EA. Centrosome duplication: of rules and licenses. Trends in cell biology. 2007;17:215–221. doi: 10.1016/j.tcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 11.Fukasawa K. Centrosome amplification, chromosome instability and cancer development. Cancer letters. 2005;230:6–19. doi: 10.1016/j.canlet.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 12.Salisbury JL. Centrosomes: coiled-coils organize the cell center. Current biology: CB. 2003;13:R88–R90. doi: 10.1016/s0960-9822(03)00033-2. [DOI] [PubMed] [Google Scholar]
- 13.Mardin BR, Schiebel E. Breaking the ties that bind: new advances in centrosome biology. J Cell Biol. 2012;197:11–18. doi: 10.1083/jcb.201108006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nigg EA, Stearns T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol. 2011;13:1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang G, et al. The role of mitotic kinases in coupling the centrosome cycle with the assembly of the mitotic spindle. J Cell Sci. 2014 doi: 10.1242/jcs.151753. [DOI] [PubMed] [Google Scholar]
- 16.Gonczy P. Towards a molecular architecture of centriole assembly. Nat Rev Mol Cell Biol. 2012;13:425–435. doi: 10.1038/nrm3373. [DOI] [PubMed] [Google Scholar]
- 17.Wong C, Stearns T. Centrosome number is controlled by a centrosome-intrinsic block to reduplication. Nature cell biology. 2003;5:539–544. doi: 10.1038/ncb993. [DOI] [PubMed] [Google Scholar]
- 18.Tsou MF, et al. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell. 2009;17:344–354. doi: 10.1016/j.devcel.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schockel L, et al. Cleavage of cohesin rings coordinates the separation of centrioles and chromatids. Nat Cell Biol. 2011;13:966–972. doi: 10.1038/ncb2280. [DOI] [PubMed] [Google Scholar]
- 20.Oliveira RA, Nasmyth K. Cohesin cleavage is insufficient for centriole disengagement in Drosophila. Current biology: CB. 2013;23:R601–R603. doi: 10.1016/j.cub.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Cabral G, et al. Multiple mechanisms contribute to centriole separation in C. elegans. Current biology : CB. 2013;23:1380–1387. doi: 10.1016/j.cub.2013.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, et al. sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by Plk1. Developmental cell. 2008;14:331–341. doi: 10.1016/j.devcel.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thein KH, et al. Astrin is required for the maintenance of sister chromatid cohesion and centrosome integrity. The Journal of cell biology. 2007;178:345–354. doi: 10.1083/jcb.200701163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura A, et al. Centrosomal Aki1 and cohesin function in separase-regulated centriole disengagement. The Journal of cell biology. 2009;187:607–614. doi: 10.1083/jcb.200906019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nigg EA, Stearns T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nature cell biology. 2011;13:1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strnad P, Gonczy P. Mechanisms of procentriole formation. Trends in cell biology. 2008;18:389–396. doi: 10.1016/j.tcb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 27.Bettencourt-Dias M, et al. SAK/PLK4 is required for centriole duplication and flagella development. Current biology : CB. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 28.Habedanck R, et al. The Polo kinase Plk4 functions in centriole duplication. Nature cell biology. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- 29.Holland AJ, et al. Centriole duplication: A lesson in self-control. Cell Cycle. 2010;9:2731–2736. doi: 10.4161/cc.9.14.12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu L, et al. Downregulation of polo-like kinase 4 in hepatocellular carcinoma associates with poor prognosis. PloS one. 2012;7:e41293. doi: 10.1371/journal.pone.0041293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ko MA, et al. Plk4 haploinsufficiency causes mitotic infidelity and carcinogenesis. Nat Genet. 2005;37:883–888. doi: 10.1038/ng1605. [DOI] [PubMed] [Google Scholar]
- 32.van Breugel M, et al. Structures of SAS-6 suggest its organization in centrioles. Science. 2011;331:1196–1199. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]
- 33.Strnad P, et al. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Developmental cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leidel S, et al. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nature cell biology. 2005;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- 35.Puklowski A, et al. The SCF-FBXW5 E3-ubiquitin ligase is regulated by PLK4 and targets HsSAS-6 to control centrosome duplication. Nature cell biology. 2011;13:1004–1009. doi: 10.1038/ncb2282. [DOI] [PubMed] [Google Scholar]
- 36.Tang CJ, et al. CPAP is a cell-cycle regulated protein that controls centriole length. Nature cell biology. 2009;11:825–831. doi: 10.1038/ncb1889. [DOI] [PubMed] [Google Scholar]
- 37.Hsu WB, et al. Functional characterization of the microtubule-binding and - destabilizing domains of CPAP and d-SAS-4. Experimental cell research. 2008;314:2591–2602. doi: 10.1016/j.yexcr.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 38.D'Angiolella V, et al. SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138–142. doi: 10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiese C, Zheng Y. Microtubule nucleation: gamma-tubulin and beyond. Journal of cell science. 2006;119:4143–4153. doi: 10.1242/jcs.03226. [DOI] [PubMed] [Google Scholar]
- 40.Lane HA, Nigg EA. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J Cell Biol. 1996;135:1701–1713. doi: 10.1083/jcb.135.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hannak E, et al. Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans. The Journal of cell biology. 2001;155:1109–1116. doi: 10.1083/jcb.200108051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lens SM, et al. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nature reviews. Cancer. 2010;10:825–841. doi: 10.1038/nrc2964. [DOI] [PubMed] [Google Scholar]
- 43.Eyers PA, et al. A novel mechanism for activation of the protein kinase Aurora A. Current biology : CB. 2003;13:691–697. doi: 10.1016/s0960-9822(03)00166-0. [DOI] [PubMed] [Google Scholar]
- 44.Tsai MY, et al. A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nature cell biology. 2003;5:242–248. doi: 10.1038/ncb936. [DOI] [PubMed] [Google Scholar]
- 45.De Luca M, et al. A functional interplay between Aurora-A, Plk1 and TPX2 at spindle poles: Plk1 controls centrosomal localization of Aurora-A and TPX2 spindle association. Cell cycle. 2006;5:296–303. doi: 10.4161/cc.5.3.2392. [DOI] [PubMed] [Google Scholar]
- 46.Santamaria A, et al. The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.004457. M110 004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee K, Rhee K. PLK1 phosphorylation of pericentrin initiates centrosome maturation at the onset of mitosis. The Journal of cell biology. 2011;195:1093–1101. doi: 10.1083/jcb.201106093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu H, et al. FAM29A promotes microtubule amplification via recruitment of the NEDD1-gamma-tubulin complex to the mitotic spindle. The Journal of cell biology. 2008;183:835–848. doi: 10.1083/jcb.200807046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mayor T, et al. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J Cell Biol. 2000;151:837–846. doi: 10.1083/jcb.151.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fry AM. The Nek2 protein kinase: a novel regulator of centrosome structure. Oncogene. 2002;21:6184–6194. doi: 10.1038/sj.onc.1205711. [DOI] [PubMed] [Google Scholar]
- 51.Faragher AJ, Fry AM. Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Molecular biology of the cell. 2003;14:2876–2889. doi: 10.1091/mbc.E03-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mardin BR, et al. Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nature cell biology. 2010;12:1166–1176. doi: 10.1038/ncb2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jackman M, et al. Human cyclins B1 and B2 are localized to strikingly different structures: B1 to microtubules, B2 primarily to the Golgi apparatus. EMBO J. 1995;14:1646–1654. doi: 10.1002/j.1460-2075.1995.tb07153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spalluto C, et al. Evidence for centriolar satellite localization of CDK1 and cyclin B2. Cell Cycle. 2013;12:1802–1803. doi: 10.4161/cc.24840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshitome S, et al. The C-terminal seven amino acids in the cytoplasmic retention signal region of cyclin B2 are required for normal bipolar spindle formation in Xenopus oocytes and embryos. Mol Cancer Res. 2003;1:589–597. [PubMed] [Google Scholar]
- 56.Macurek L, et al. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 2008;455:119–123. doi: 10.1038/nature07185. [DOI] [PubMed] [Google Scholar]
- 57.Mardin BR, et al. Plk1 controls the Nek2A-PP1gamma antagonism in centrosome disjunction. Curr Biol. 2011;21:1145–1151. doi: 10.1016/j.cub.2011.05.047. [DOI] [PubMed] [Google Scholar]
- 58.Wu CC, et al. p53 negatively regulates Aurora A via both transcriptional and posttranslational regulation. Cell cycle (Georgetown, Tex.) 2012;11:3433–3442. doi: 10.4161/cc.21732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaplan DD, et al. Identification of a role for beta-catenin in the establishment of a bipolar mitotic spindle. The Journal of biological chemistry. 2004;279:10829–10832. doi: 10.1074/jbc.C400035200. [DOI] [PubMed] [Google Scholar]
- 60.Bahmanyar S, et al. beta-Catenin is a Nek2 substrate involved in centrosome separation. Genes & development. 2008;22:91–105. doi: 10.1101/gad.1596308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Graser S, et al. Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. Journal of cell science. 2007;120:4321–4331. doi: 10.1242/jcs.020248. [DOI] [PubMed] [Google Scholar]
- 62.Tanenbaum ME, Medema RH. Mechanisms of centrosome separation and bipolar spindle assembly. Dev Cell. 2010;19:797–806. doi: 10.1016/j.devcel.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 63.Bertran MT, et al. Nek9 is a Plk1-activated kinase that controls early centrosome separation through Nek6/7 and Eg5. The EMBO journal. 2011;30:2634–2647. doi: 10.1038/emboj.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cahu J, et al. Phosphorylation by Cdk1 increases the binding of Eg5 to microtubules in vitro and in Xenopus egg extract spindles. PLoS One. 2008;3:e3936. doi: 10.1371/journal.pone.0003936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gavet O, Pines J. Activation of cyclin B1-Cdk1 synchronizes events in the nucleus and the cytoplasm at mitosis. J Cell Biol. 2010;189:247–259. doi: 10.1083/jcb.200909144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCleland ML, O'Farrell PH. RNAi of mitotic cyclins in Drosophila uncouples the nuclear and centrosome cycle. Curr Biol. 2008;18:245–254. doi: 10.1016/j.cub.2008.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gayek AS, Ohi R. Kinetochore-microtubule stability governs the metaphase requirement for Eg5. Molecular biology of the cell. 2014;25:2051–2060. doi: 10.1091/mbc.E14-03-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tanenbaum ME, et al. Dynein, Lis1 and CLIP-170 counteract Eg5-dependent centrosome separation during bipolar spindle assembly. The EMBO journal. 2008;27:3235–3245. doi: 10.1038/emboj.2008.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Raaijmakers JA, et al. Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. EMBO J. 2012;31:4179–4190. doi: 10.1038/emboj.2012.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Toso A, et al. Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. The Journal of cell biology. 2009;184:365–372. doi: 10.1083/jcb.200809055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khodjakov A, et al. Centrosome-independent mitotic spindle formation in vertebrates. Current biology: CB. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- 72.Hinchcliffe EH, et al. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science. 2001;291:1547–1550. doi: 10.1126/science.1056866. [DOI] [PubMed] [Google Scholar]
- 73.Rosenblatt J, et al. Myosin II-dependent cortical movement is required for centrosome separation and positioning during mitotic spindle assembly. Cell. 2004;117:361–372. doi: 10.1016/s0092-8674(04)00341-1. [DOI] [PubMed] [Google Scholar]
- 74.Toso A, et al. Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. The Journal of Cell Biology. 2009;184:365–372. doi: 10.1083/jcb.200809055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rosenblatt J, et al. Myosin II-Dependent Cortical Movement Is Required for Centrosome Separation and Positioning during Mitotic Spindle Assembly. Cell. 2004;117:361–372. doi: 10.1016/s0092-8674(04)00341-1. [DOI] [PubMed] [Google Scholar]
- 76.Basto R, et al. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 77.Silkworth WT, et al. Timing of centrosome separation is important for accurate chromosome segregation. Molecular Biology of the Cell. 2012;23:401–411. doi: 10.1091/mbc.E11-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Silkworth WT, Cimini D. Transient defects of mitotic spindle geometry and chromosome segregation errors. Cell division. 2012;7:19. doi: 10.1186/1747-1028-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lampson MA, Cheeseman IM. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends in cell biology. 2011;21:133–140. doi: 10.1016/j.tcb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cimini D, et al. Aurora Kinase Promotes Turnover of Kinetochore Microtubules to Reduce Chromosome Segregation Errors. Current Biology. 2006;16:1711–1718. doi: 10.1016/j.cub.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 81.Tanaka TU, et al. Evidence that the Ipl1-Sli15 (Aurora Kinase-INCENP) Complex Promotes Chromosome Bi-orientation by Altering Kinetochore-Spindle Pole Connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 82.Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. The Journal of Cell Biology. 2009;184:373–381. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Uchida KSK, et al. Kinetochore stretching inactivates the spindle assembly checkpoint. The Journal of Cell Biology. 2009;184:383–390. doi: 10.1083/jcb.200811028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Castillo A, et al. Overexpression of Eg5 causes genomic instability and tumor formation in mice. Cancer Res. 2007;67:10138–10147. doi: 10.1158/0008-5472.CAN-07-0326. [DOI] [PubMed] [Google Scholar]
- 85.Lu MS, Johnston CA. Molecular pathways regulating mitotic spindle orientation in animal cells. Development. 2013;140:1843–1856. doi: 10.1242/dev.087627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee CY, et al. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature. 2006;439:594–598. doi: 10.1038/nature04299. [DOI] [PubMed] [Google Scholar]
- 87.Inaba M, Yamashita YM. Asymmetric stem cell division: precision for robustness. Cell stem cell. 2012;11:461–469. doi: 10.1016/j.stem.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 88.Bergstralh DT, St Johnston D. Spindle orientation: What if it goes wrong? Seminars in cell & developmental biology. 2014 doi: 10.1016/j.semcdb.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Du Q, Macara IG. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell. 2004;119:503–516. doi: 10.1016/j.cell.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 90.Thery M, et al. The extracellular matrix guides the orientation of the cell division axis. Nature cell biology. 2005;7:947–953. doi: 10.1038/ncb1307. [DOI] [PubMed] [Google Scholar]
- 91.Thery M, et al. Experimental and theoretical study of mitotic spindle orientation. Nature. 2007;447:493–496. doi: 10.1038/nature05786. [DOI] [PubMed] [Google Scholar]
- 92.Fink J, et al. External forces control mitotic spindle positioning. Nature cell biology. 2011;13:771–778. doi: 10.1038/ncb2269. [DOI] [PubMed] [Google Scholar]
- 93.Smith E, et al. Differential control of Eg5-dependent centrosome separation by Plk1 and Cdk1. EMBO J. 2011;30:2233–2245. doi: 10.1038/emboj.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kotak S, et al. Cortical dynein is critical for proper spindle positioning in human cells. The Journal of cell biology. 2012;199:97–110. doi: 10.1083/jcb.201203166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van Heesbeen RG, et al. Nuclear envelope-associated dynein cooperates with Eg5 to drive prophase centrosome separation. Communicative & integrative biology. 2013;6:e23841. doi: 10.4161/cib.23841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Carter SL, et al. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]