Abstract
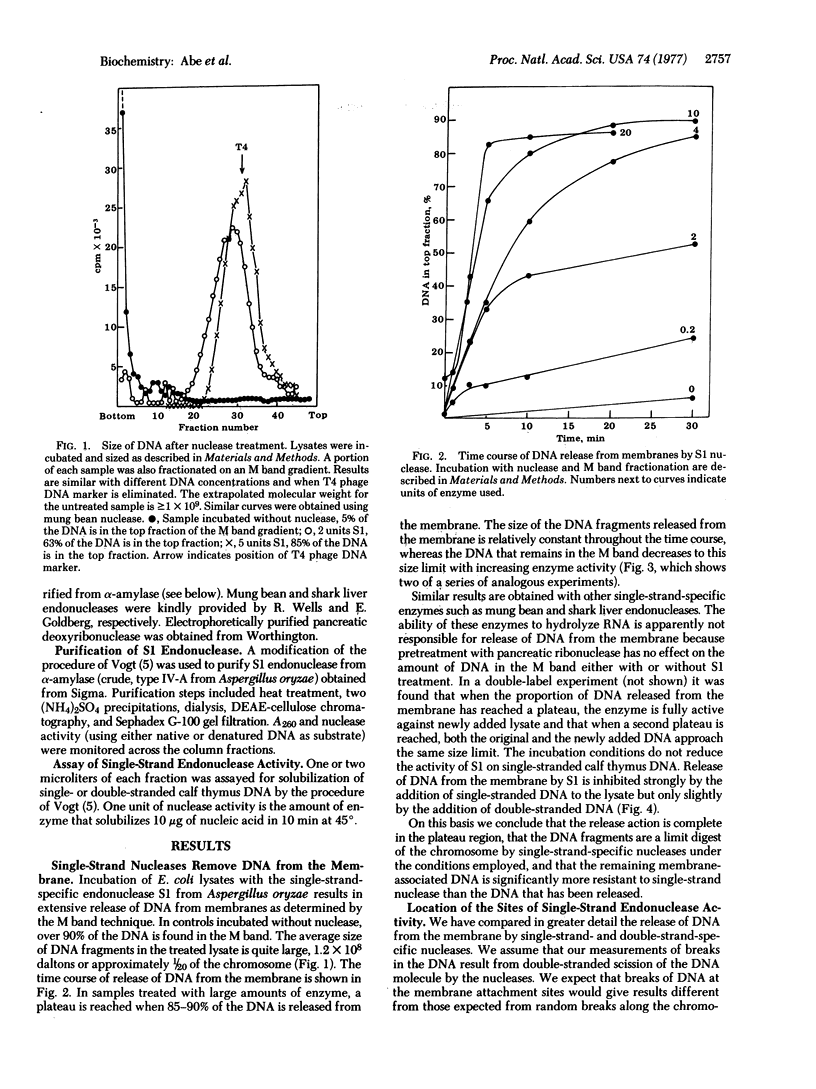
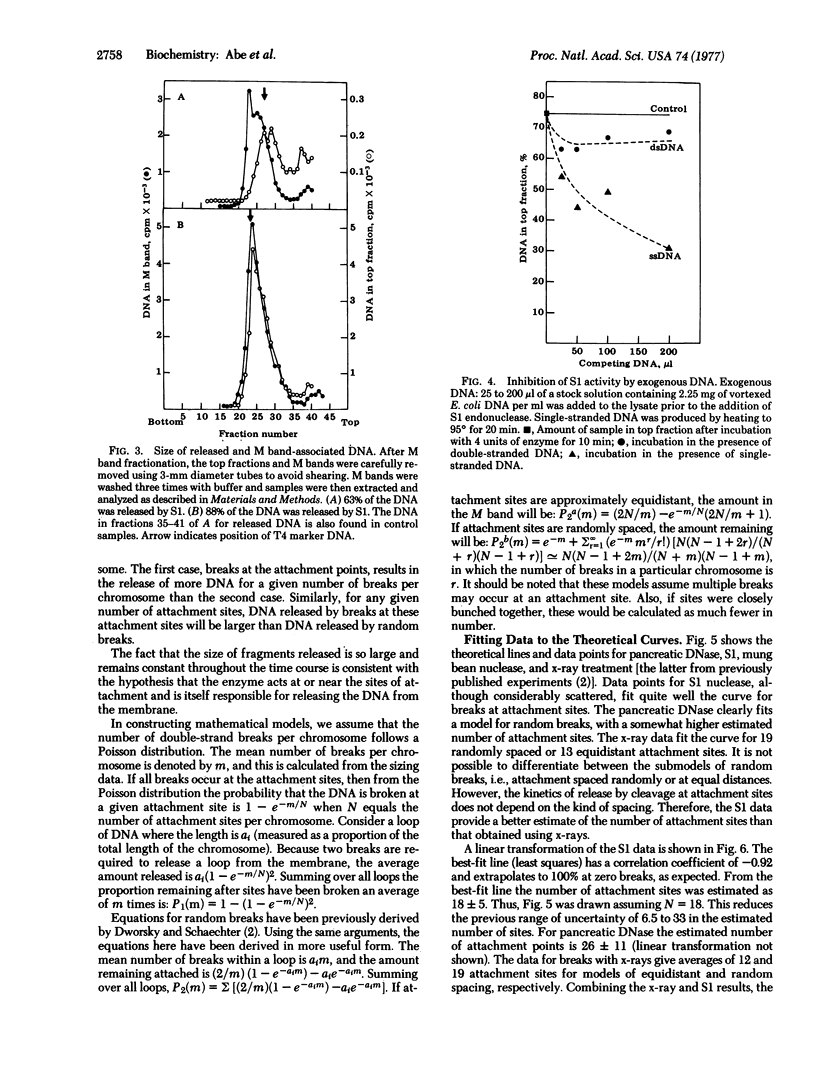
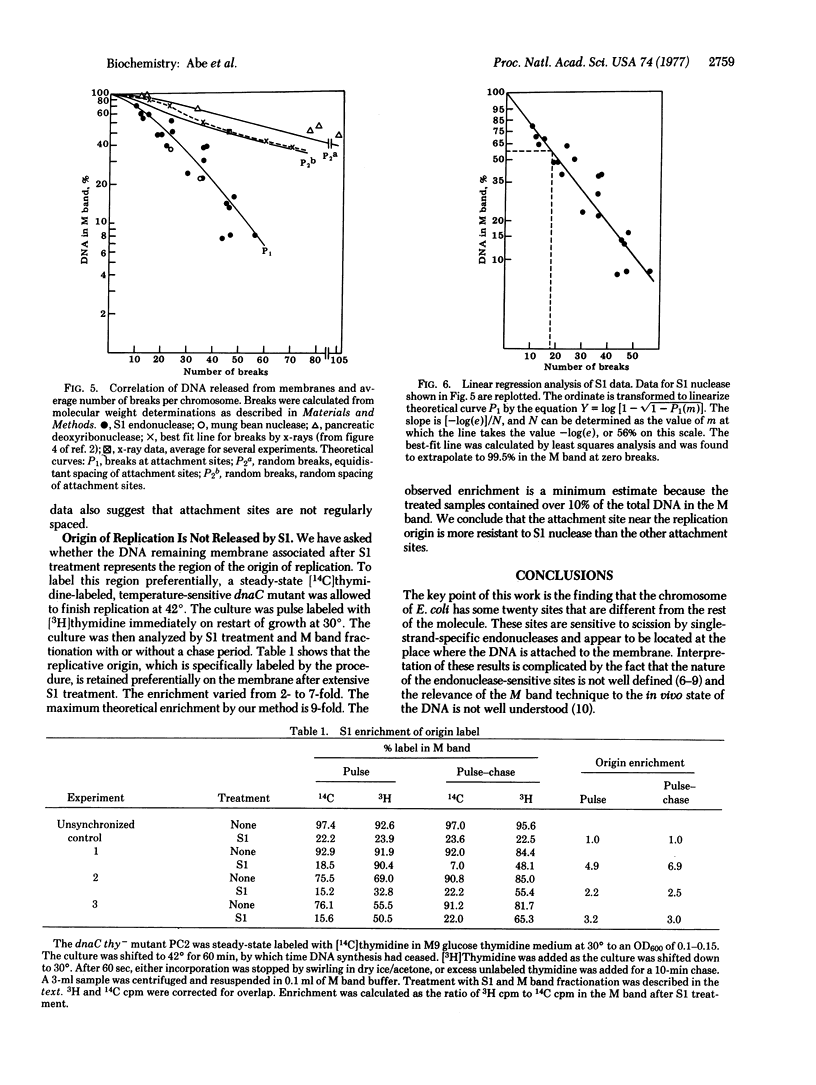
Treatment of gently prepared lysates of Escherichia coli with single-strand-specific endonuclease (SI or from mung beans) results in the release of about 90% of the DNA from membranes, as determined by the M band technique. The released DNA has an average molecular weight of about 1.2 X 10(8). Data obtained with endonuclease S1 fit a mathematical model in which substrate sites are at or near membrane attachment sites. Data obtained with pancreatic deoxyribonuclease or x-rays fit a model for double-strand breaks at random sites along the DNA. Fitting data to these models, we estimate that there are 18+/-5 membrane attachment sites. The DNA remaining after S1 nuclease treatment is enriched for the region near the origin of chromosome replication. Therefore, attachment at this region near the origin of chromosome replication. Therefore, attachment at this region appears to be chemically different from that at the other sites along the DNA.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chan H. W., Wells R. D. Structural uniqueness of lactose operator. Nature. 1974 Nov 15;252(5480):205–209. doi: 10.1038/252205a0. [DOI] [PubMed] [Google Scholar]
- Chowdhury K., Sauer G. S1 nuclease converts relaxed circular closed duplex DNA to open linear DNA of unit length. FEBS Lett. 1976 Sep 15;68(1):68–70. doi: 10.1016/0014-5793(76)80406-1. [DOI] [PubMed] [Google Scholar]
- Dworsky P., Schaechter M. Effect of rifampin on the structure and membrane attachment of the nucleoid of Escherichia coli. J Bacteriol. 1973 Dec;116(3):1364–1374. doi: 10.1128/jb.116.3.1364-1374.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakan S., Turner G. N., Pagano J. S., Hancock R. Sites of replication of chromosomal DNA in a eukaryotic cell. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2300–2305. doi: 10.1073/pnas.69.8.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freifelder D. Molecular weights of coliphages and coliphage DNA. IV. Molecular weights of DNA from bacteriophages T4, T5 and T7 and the general problem of determination of M. J Mol Biol. 1970 Dec 28;54(3):567–577. doi: 10.1016/0022-2836(70)90127-0. [DOI] [PubMed] [Google Scholar]
- Leibowitz P. J., Schaechter M. The attachment of the bacterial chromosome to the cell membrane. Int Rev Cytol. 1975;41:1–28. doi: 10.1016/s0074-7696(08)60964-x. [DOI] [PubMed] [Google Scholar]
- Rosenberg B. H., Cavalieri L. F. Shear sensitivity of the E. coli genome: multiple membrane attachment points of the E. coli DNA. Cold Spring Harb Symp Quant Biol. 1968;33:65–72. doi: 10.1101/sqb.1968.033.01.012. [DOI] [PubMed] [Google Scholar]
- Vogt V. M. Purification and further properties of single-strand-specific nuclease from Aspergillus oryzae. Eur J Biochem. 1973 Feb 15;33(1):192–200. doi: 10.1111/j.1432-1033.1973.tb02669.x. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Interactions between twisted DNAs and enzymes: the effects of superhelical turns. J Mol Biol. 1974 Aug 25;87(4):797–816. doi: 10.1016/0022-2836(74)90085-0. [DOI] [PubMed] [Google Scholar]
- Wiegand R. C., Godson G. N., Radding C. M. Specificity of the S1 nuclease from Aspergillus oryzae. J Biol Chem. 1975 Nov 25;250(22):8848–8855. [PubMed] [Google Scholar]
- Worcel A., Burgi E. On the structure of the folded chromosome of Escherichia coli. J Mol Biol. 1972 Nov 14;71(2):127–147. doi: 10.1016/0022-2836(72)90342-7. [DOI] [PubMed] [Google Scholar]


