Table 2.
Amino Acid Substrate Specificity of wt TH and Selected Mutants
| Protein (mutation) | Substrate | Vmax (nmol min−1mg−1) | Km (μM) | 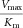 |
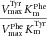 |
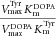 |
|---|---|---|---|---|---|---|
| wt TH | l-Tyr | 2460 ± 417 | 5.1 ± 2.2 | 484 | 88 | 114 |
| l-Phe | 1640 ± 148 | 296 ± 69 | 5.5 | |||
| l-Dopa | 126 ± 11 | 29.8 ± 8.5 | 4.2 | |||
| p.Arg233His | l-Tyr | 150 ± 30 | 13.0 ± 4.0 | 11.5 | 4.8 | 7.7 |
| (c.698A>G) | l-Phe | 352 ± 33 | 149 ± 22 | 2.4 | ||
| l-Dopa | 1.6 | 1.0 | 1.5 | |||
| p.Gly247Ser | l-Tyr | 1189 ± 53 | 11.8 ± 0.9 | 101 | 16 | 36 |
| (c.739G>A) | l-Phe | 734 ± 46 | 119 ± 25 | 6.2 | ||
| l-Dopa | 50 ± 9.4 | 17 ± 12 | 2.8 | |||
| p.Cys359Phe | l-Tyr | 329.4 ± 4.2 | 37.6 ± 1.1 | 8.8 | 80 | |
| (c.1076G>T) | l-Phe | 73.2 ± 5.7 | 663 ± 99 | 0.1 | ||
| l-Dopa | n.m. | n.m. | ||||
| p.Phe375Leu | l-Tyr | 241 ± 20 | 56 ± 11 | 4.3 | 2.2 | 11 |
| (c.1125C>G) | l-Phe | 1960 ± 44 | 993 ± 37 | 2.0 | ||
| l-Dopa | 77 ± 30 | 202 ± 135 | 0.4 | |||
| p.Leu387Met | l-Tyr | 2830 ± 1450 | 24 ± 17 | 118 | 21 | 131 |
| (c.1159C>A) | l-Phe | 1407 ± 78 | 258 ± 38 | 5.5 | ||
| l-Dopa | 62 ± 14 | 68 ± 39 | 0.9 | |||
| p.Gly414Arg | l-Tyr | 66.8 ± 7.5 | 26.6 ± 8.1 | 2.5 | 0.5 | 1.3 |
| (c.1240G>A) | l-Phe | 452 ± 17 | 92 ± 12 | 4.9 | ||
| l-Dopa | 35.1 ± 7.2 | 18 ± 13 | 1.9 |
The table summarizes the different enzyme kinetic parameters obtained for wt TH and some of the mutants, using l-Tyr, l-Phe, or l-Dopa as substrate. We did not observe substrate inhibition kinetics for l-Phe or l-Dopa, which is why only Vmax and Km values are reported. The data for l-Tyr are taken from Table1. Data points were fitted using standard MM kinetics by nonlinear regression (Sigma plot) as described in Materials and Methods section. Values are given as the best estimate ± standard error of estimate. n.m., not measured.
