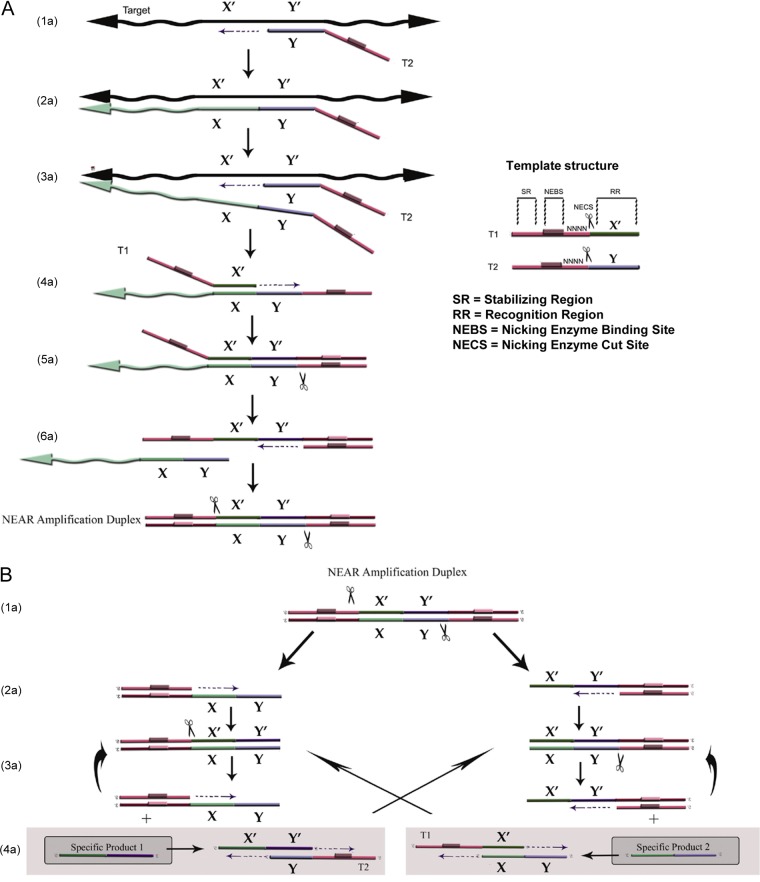
FIG 1.
NEAR mechanism. (A) Mechanism of NEAR amplification duplex formation. (1a and 2a) The recognition region of T2 binds to the complementary target region and is extended by polymerase along the target. (3a) A second T2 binds to the same target and is extended, displacing the first T2. (4a) The recognition region of T1 binds to its complement in the released strand and is extended to the 5′ end, creating a double-stranded nicking enzyme recognition site. (5a) Nicking enzyme binds and nicks (indicated by scissors). (6a) polymerase synthesizes off the cleaved 3′ OH along T1, displacing the remaining target complement, and the final extended double-stranded complex is termed the NEAR amplification duplex. (B) Mechanism of product formation. (1b and 2b) Nicking enzymes bind to both nicking enzyme recognition sites on the NEAR duplex; cleavage and strand displacement amplification at both sites creates two complexes, each consisting of a duplex stability region, a nicking enzyme recognition region, and a single-stranded target. (3b and 4b) Repeated nicking, polymerization, and strand displacement result in the amplification of products 1 and 2. Cleaved complexes are regenerated (3b), while products 1 and 2 can anneal to T1 and T2, respectively (4b), resulting in bidirectional extension and creating duplexes that generate the opposite product upon cleavage. The products continue to recycle until the templates, deoxynucleoside triphosphates (dNTPs), or enzymes are depleted.