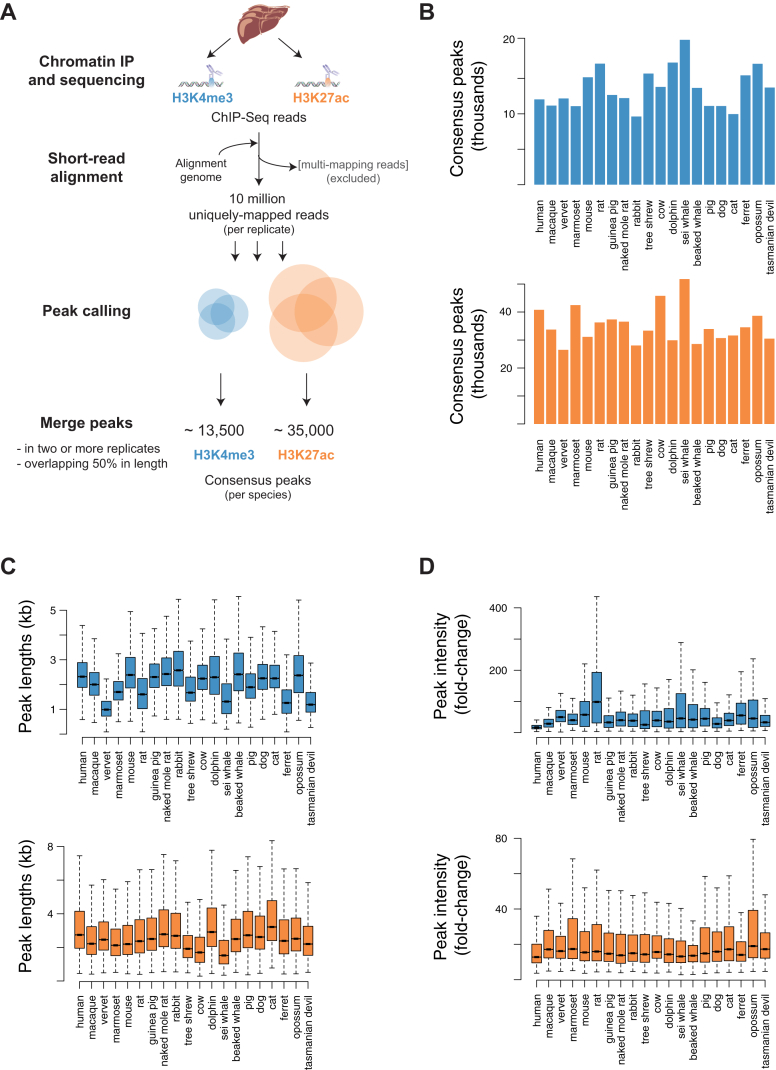
Figure S1.
Analysis Workflow and Quality Control of H3K4me3 and H3K27ac ChIP-Seq in 20 Mammals, Related to Figure 1
(A) Short-read alignment and peak calling workflow (see also Extended Experimental Procedures)
(B) Numbers of consensus peaks identified for H3K4me3 (blue) or H3K27ac (orange) in each species’ liver tissue.
(C) Length distributions of consensus H3K4me3 (blue) or H3K27ac (orange) peaks are represented as boxplots for each species.
(D) Peak intensity distributions are represented as boxplots for each species’ data (H3K4me3, blue; H3K27ac, orange). Peak intensities correspond to average fold enrichment values over total input DNA across biological replicates (see Extended Experimental Procedures).