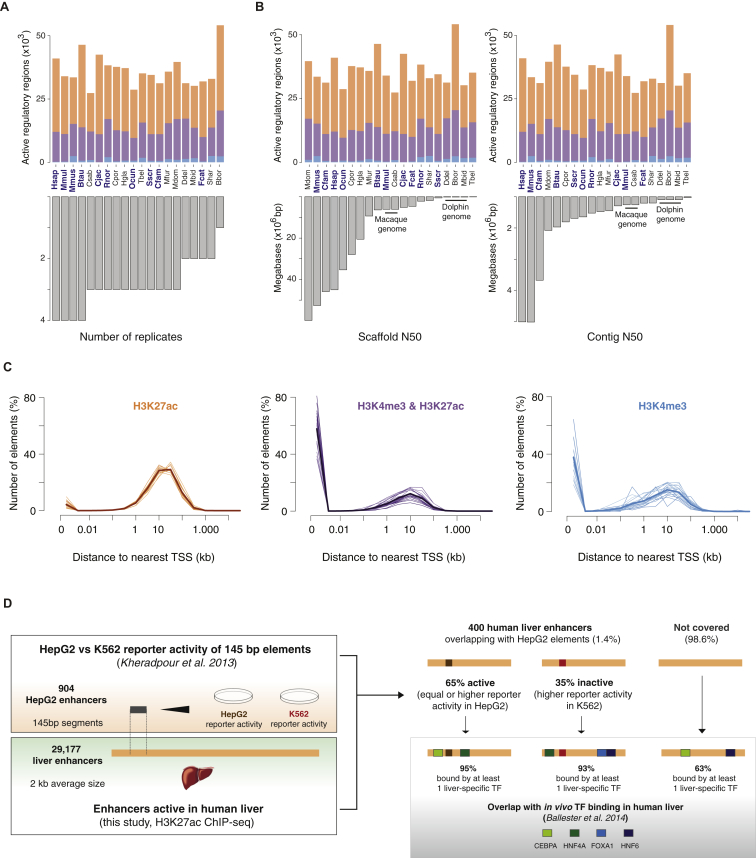
Figure S2.
Quality Control of Experimental Promoter and Enhancer Definition, Related to Figure 1
(A) Numbers of experimentally identified promoters (H3K4me3&H3K27ac, purple; H3K4me3, blue) and enhancers (H3K27ac, orange) per species are represented as stacked barplots in the upper plot, ordered by decreasing number of biological replicates used for each species (lower plot). Except for Bbor (Balaenoptera borealis), where a single replicate was used, the number of biological replicates has little influence on the number of active regulatory regions identified per species.
(B) As in (A), but numbers of promoters/enhancers in each species are now ordered by decreasing scaffold or contig N50 values, both indicative of genome assembly quality. Species highlighted in blue correspond to genomes in the EPO multiple alignment, considered to be the highest-quality reference genomes. Assembly qualities do not appear to influence experimental variation in the number of promoters or enhancers identified in each species.
(C) The distribution of distances to the nearest transcriptional start site (TSS) was calculated for all experimentally identified regions in each species’ data (thin lines). Bolded lines represent the average distance distribution across all species for H3K4me3 (blue), H3K4me3&H3K27ac (purple) and H3K27ac (orange) elements. In agreement with their categorisation as enhancer elements, in all species most H3K27ac locations are distal to coding regions. Both H3K4me3 and H3K4me3&H3K27ac elements are largely located close to annotated TSSs consistent with being proximal promoters. The minority of distal elements marked by H3K4me3 or H3K4me3&H3K27ac may correspond to unannotated transcripts; further, the latter may also act as enhancers (Kim et al., 2010).
(D) H3K27ac-defined enhancers enrich for regulatory activity: Human liver enhancers identified in this study through H3K27ac ChIP-seq (bottom inset) were overlapped with 145 bp sequence elements assayed for reporter activity in human liver carcinoma (HepG2) and human erythroleukemia cells (K562) (top inset; Kheradpour et al., 2013). These correspond to enhancer candidates identified in HepG2 cells and containing motifs for liver-specific transcription factors.
Four hundred human liver enhancers contained at least one 145 bp segment (1.1 segments per enhancer on average). 65% of these enhancers were active based on the reporter activity of the assayed segments, which displayed higher activity in HepG2 compared to K562 cells, or equal activity in both cell lines. The remaining 35% human liver enhancers overlapped segments having higher activity in K562 cells, and were thus classified as inactive in HepG2 cells. Grey inset: Human liver enhancers identified in this study were overlapped with in vivo binding locations for four liver-specific transcription factors, as reported independently in human liver samples (Ballester et al., 2014). Among the 400 enhancers containing segments assayed in Kheradpour et al., 93%–95% of them were bound by at least one liver-specific TF, regardless of the reporter activity of their overlapping segments. This suggests that in cases where the overlapping segment was inactive in the reporter assay, the corresponding enhancer may harbor regulatory activity outside the interrogated sequence. Across all liver enhancers in human, 63% are bound by at least one of the four liver-specific transcription factors, in line with previous estimates of functional enhancer activity in H3K27ac-marked regions (Nord et al., 2013).