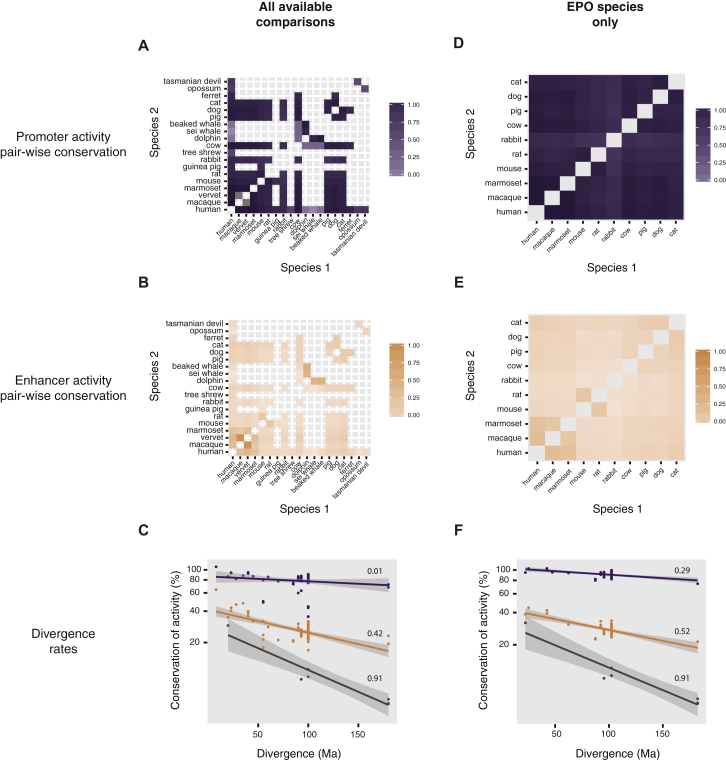
Figure S4.
Experimentally Determined Rates of Promoter and Enhancer Evolution across Mammals, Related to Figure 4
(A–F) The average fraction of regions with pairwise conserved activity for promoters (purple) or enhancers (orange) represented as heatmaps, as measured for: (1) all available comparisons in the dataset (A and B), using the 13 eutherian mammals Ensembl EPO multiple alignment where possible and ad hoc LastZ pairwise alignments otherwise (see Extended Experimental Procedures). (2) only species in the 13 eutherian mammals Ensembl.
EPO multiple alignment (D and E), corresponding to the higher-quality reference genomes in the dataset. The choice of species and alignments had no significant influence on the rates, as calculated by an exponential decay fit to either set of comparisons. Percent conservation (y axis) is shown in logarithmic scale, and numbers above each dataset represent R2 values for the exponential decay fit (C and F). The regressions (solid lines) in (C) were used to calculate the estimated half-lives and mean lifetimes in Figure 4 (promoters: half-life 939 Ma [641-1760], mean lifetime 1355 Ma [924-2539]; enhancers: half-life 296 Ma [231-408]; mean lifetime 427 Ma [334-589]; CEBPA binding sites: half-life 144 Ma [103-237]; mean lifetime 207 Ma [148-342]). Numbers in square brackets indicate 95% confidence intervals for each value.