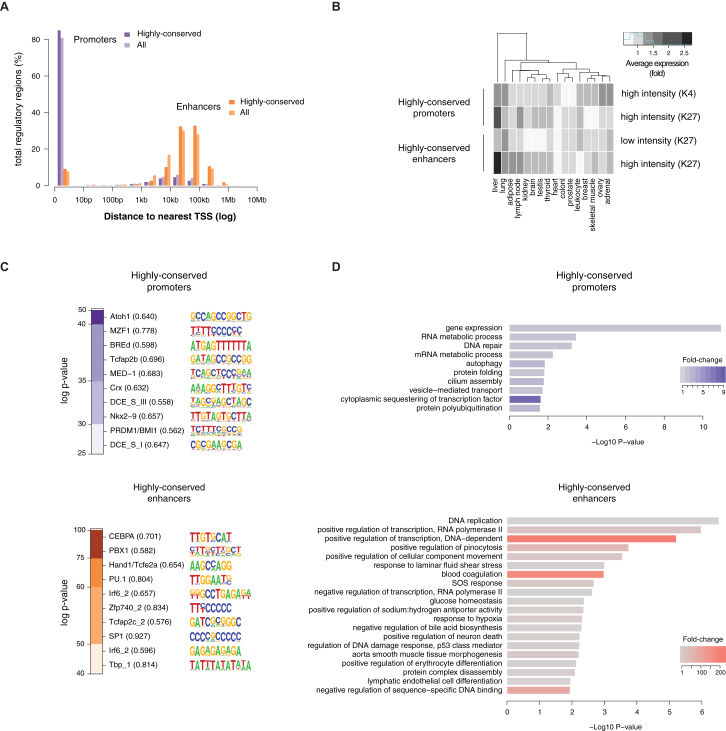
Figure S5.
Additional Properties of Highly Conserved Promoters and Enhancers, Related to Figure 5
(A) The distribution of distances to the nearest TSS is almost identical between highly conserved promoters or enhancers (darker purple and orange, respectively) and all experimentally identified promoters/enhancers in human (lighter purple and orange bars).
(B) Average expression of genes associated to highly conserved promoters and enhancers across a panel of 16 human tissues (Petryszak et al., 2014). Highly conserved enhancers are associated with genes showing a higher average expression in liver, especially for the top 50% H3K27ac intensities (“high intensity (K27)”). Conversely, highly conserved promoters are largely associated with ubiquitously expressed genes, although promoters with high H3K27ac intensity also associate with high liver gene expression. Expression profiles for genes associated with all promoters and enhancers identified in human were used as background to normalize expression values (see Extended Experimental Procedures).
(C) Sequence motifs specifically enriched in highly conserved promoters and enhancers, using all experimentally identified promoters or enhancers in human as a background control. The ten most-enriched motifs are shown, and enrichment p values are represented as heatmaps (logarithmic scale).
(D) Gene ontology annotations for biological processes enriched near highly conserved promoters and enhancers. Liver-related annotations such as blood coagulation, glucose homeostasis or bile acid biosynthesis are found for highly conserved enhancers, in line with their association to liver-specific genes.