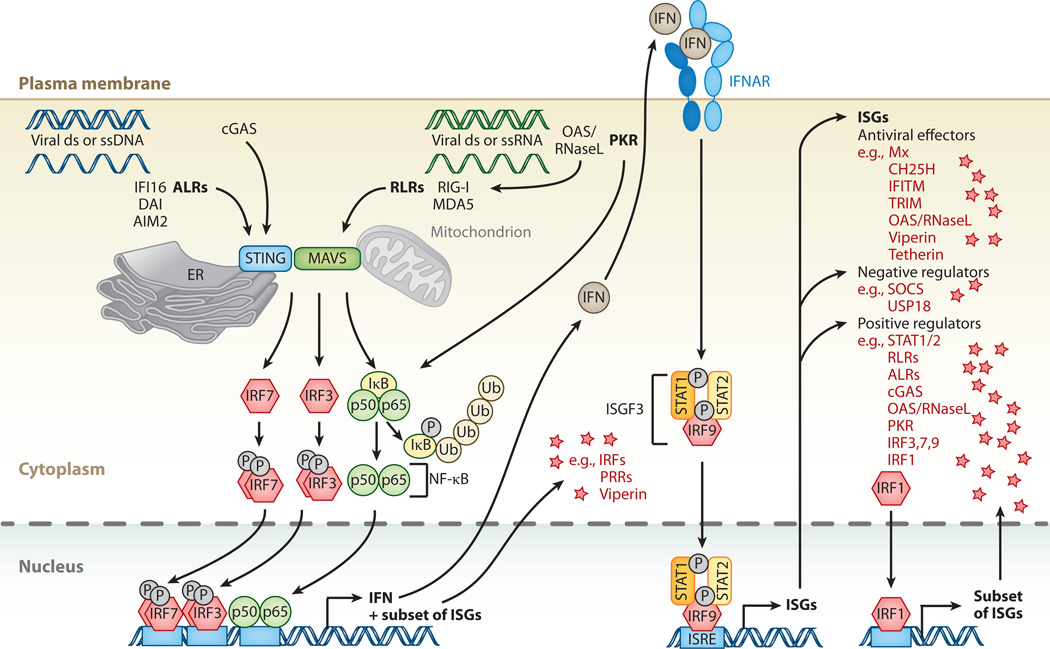
Figure 2.
Cytosolic nucleic acid pattern recognition and activation of ISGs. Cytosolic pattern-recognition receptors (PRRs) recognize viral double-stranded (ds) or single-stranded (ss) DNA or RNA. AIM2-like receptors (ALRs), such as IFI16, DAI, or AIM2 itself, specialize in DNA detection, whereas RIG-I-like receptors (RLR)—RIG-I and MDA5—specialize in RNA detection. Cyclic GMP-AMP synthase acts as an additional DNA sensor. 2′-5′-oligoadenylate synthetase (OAS) senses foreign RNA and produces 2′-5′ adenylic acid, which activates latent RNase (RNaseL). Degradation products produced by RNaseL further stimulate RLRs. Protein kinase R (PKR) is an additional sensor for foreign RNA. PRR signals are transduced to transcription factor activity by stimulator of IFN genes (STING) and mitochondrial antiviral-signaling protein (MAVS) at the ER/mitochondrion-associated membrane. Activation of STING/MAVS leads to phosphorylation of interferon (IFN) response factors 3 or 7 (IRF3/7), or to phosphorylation and ubiquitin-mediated degradation of IκB. Phosphorylated dimers of IRF3/7 or NF-κB translocate to the nucleus, where they bind to and activate specific promoters, triggering expression of IFN as well as a subset of ISGs. These ISGs include IRFs and PRRs but also antiviral effectors such as viperin. IFN induces gene expression via the JAK-STAT pathway, resulting in expression of a large spectrum of ISGs that can be divided into antiviral effectors and negative or positive regulators of IFN signaling. A special case of positive regulators is IRF1, which upon expression directly translocates to the nucleus to enhance expression of a subset of ISGs.