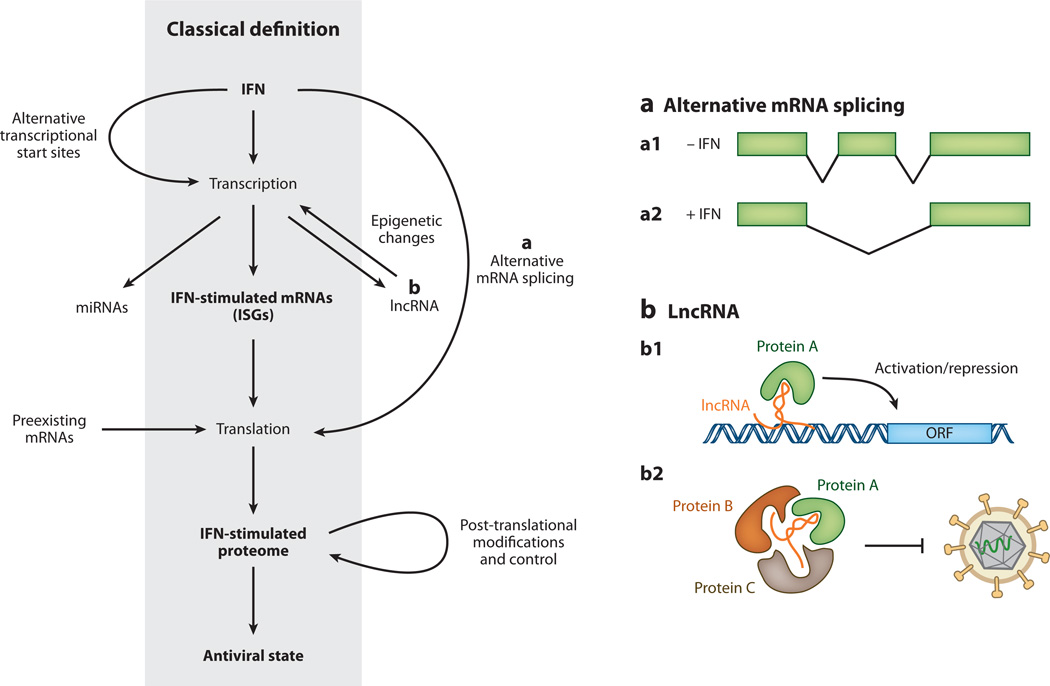
Figure 5.
Canonical and noncanonical definition of interferon (IFN)-stimulated antiviral effectors. For years the canonical understanding of the IFN-mediated antiviral response has been that IFN triggers the transcription of IFN-stimulated genes (ISGs), which leads to changes in the cellular proteome and establishment of an antiviral state within cells. Recent studies suggest that the situation is likely more complex. (a) IFN treatment of cells promotes alternative transcriptional start site usage, and IFN-induced alternative mRNA splicing may give rise to transcript isoforms that encode for different protein products or transcripts with altered stability or translational efficiency (a1/a2). (b) In addition, IFN stimulation may alter the expression of microRNAs (miRNAs) or long noncoding RNAs (lncRNAs). LncRNAs may influence gene expression through interaction with chromatin remodeling complexes (b1) or may serve as a scaffold for the formation of RNA-protein complexes that confer antiviral activity (b2). IFN stimulation or pathogen recognition may promote translation of preexisting mRNAs. Finally, IFN stimulation influences the proteome directly by promoting proteins’ post-translational modification, altering protein stability, and increasing protein secretion.