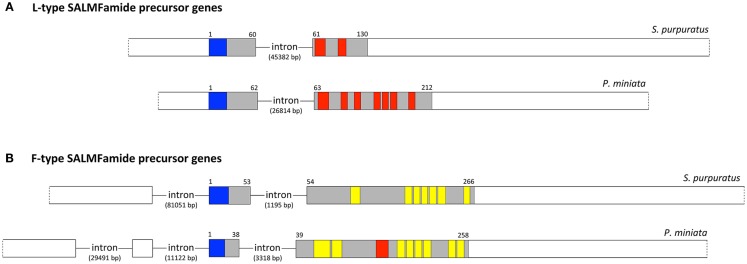
Figure 2.
Comparison of the structure of SALMFamide genes in the sea urchin S. purpuratus and the starfish P. miniata. (A). Diagrammatic representation of the structure of the L-type SALMFamide precursor gene in S. purpuratus and P. miniata. (B). Diagrammatic representation of the structure of the F-type SALMFamide precursor gene in S. purpuratus and P. miniata. Exons are drawn to scale as rectangles and introns are shown as lines but not to scale (intron lengths are shown in parentheses). Non-coding exons or regions of exons are shown in white; note that the 5′ and 3′ of the genes are shown as a dashed line because it is possible that additional non-coding bases are transcribed in vivo but are not present in transcripts that have been sequenced. The protein-coding exons or regions of exons are shaded (gray) or colored, with the N-terminal signal peptide shown in blue and the predicted neuropeptide products of the precursors shown in red (L-type SALMFamides) or yellow (F-type SALMFamides). The numbers above the protein-coding exons are amino-acid residue positions in the protein products of the genes. A feature that appears to distinguish L-type and F-type SALMFamide genes is the structure of the first protein-coding exons. Thus, in the two L-type SALMFamide genes the first protein-coding exon also encodes a 5′ non-coding region, whilst in the two F-type SALMFamide genes the first protein-coding exon does not have a 5′ non-coding region. These differences in gene structure provide important evidence that the L-type SALMFamide precursor genes in S. purpuratus and P. miniata are orthologs and the F-type SALMFamide precursor genes in S. purpuratus and P. miniata are orthologs.