Fig. 1.
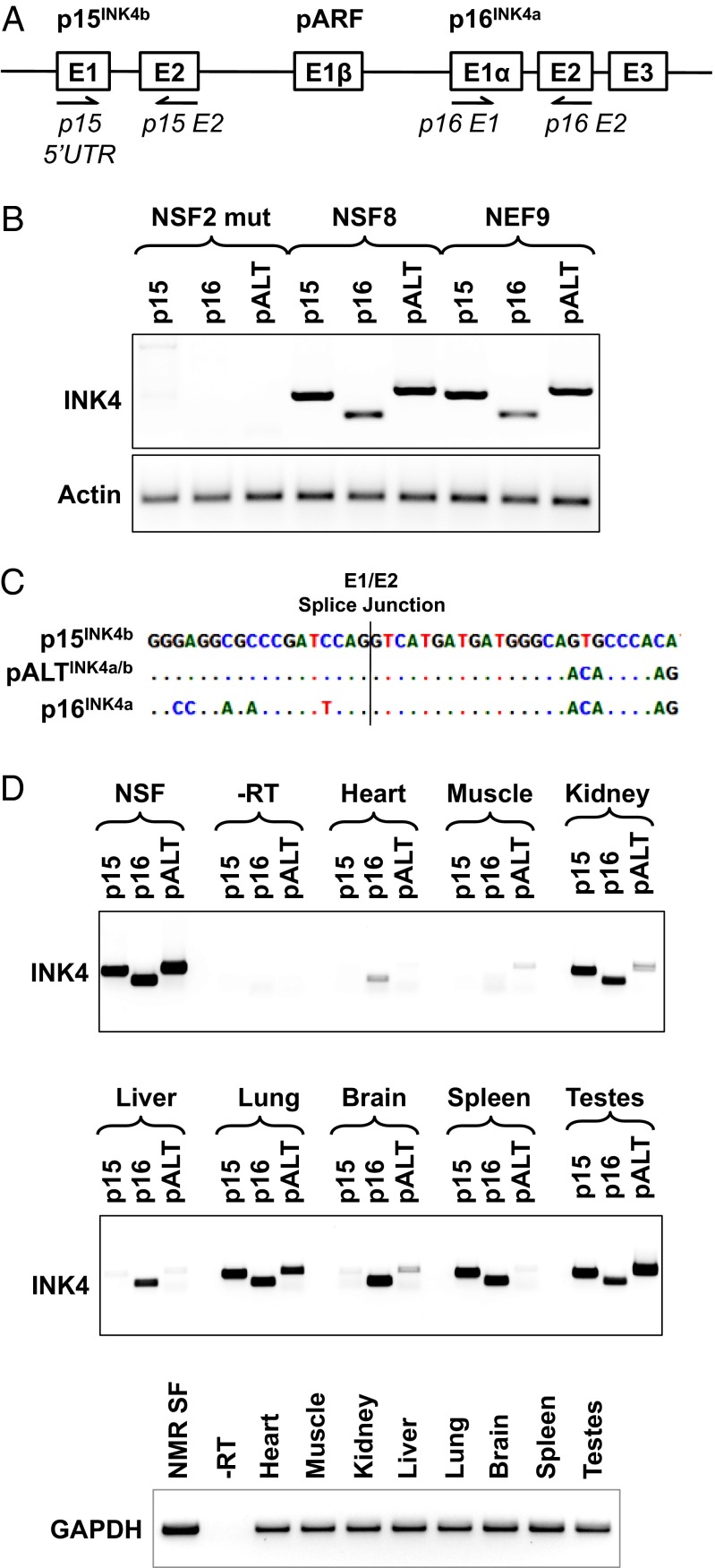
The INK4a/b locus of the NMR produces an alternative protein isoform pALTINK4a/b composed of the first exon of p15INK4b and the second and third exons of p16INK4a. (A) Schematic of the INK4a/b locus showing the location of the primers used to amplify the three INK4a/b transcripts. (B) RT-PCR products corresponding to the three INK4a/b transcripts amplified from RNA isolated from three lines of naked mole rat fibroblasts, with primers shown in A and Table S2; pALTINK4a/b was amplified with primers p15-5UTR and p16-E2. NSF2 mut is the line of spontaneously mutated naked mole rat fibroblasts that lost INK4a/b locus expression and do not show ECI; NSF8 are primary naked mole rat skin fibroblasts; NEF9 are naked mole rat embryonic fibroblasts. (C) Sequences of the Exon1/Exon2 splice junctions in p15, p16, and pALT. (D) RT-PCR showing INK4a/b products amplified from naked mole rat tissues. GAPDH was used to show that equal amounts of RNA template were used for each tissue. The reactions were repeated at least three times, and representative gels are shown.