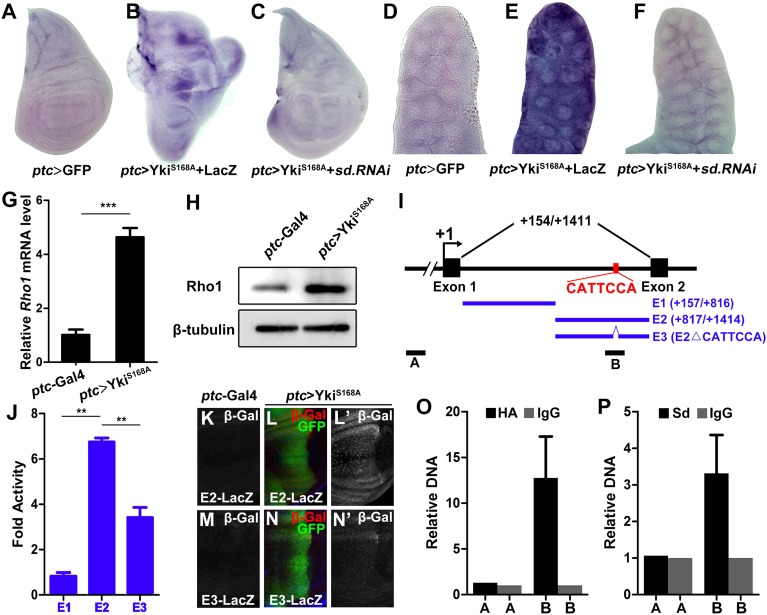
Fig. 5.
Yki–Sd directly induces Rho1 transcription. (A–F) Knocking down sd suppresses Yki-induced Rho1 transcription. In situ hybridization to Rho1 mRNA of third instar wing discs (A–C) or salivary gland (D–F) expressing GFP (A and D), YkiS168A + LacZ (B and E), YkiS168A + sd RNAi (C and F) driven by ptc–Gal4. (G and H) Ectopic Yki up-regulates Rho1 mRNA (G) and protein (H) levels, as shown by quantitative PCR and Western blot, respectively. (n = 200, mean + SD, ***P < 0.001) (I) Scheme of the Rho1 locus showing the first intron. The Sd binding motif (CATTCCA) is indicated. E1, E2, and E3 were used to drive luciferase expression; A and B represent the control and target region of ChIP assays, respectively. (J) Luciferase assay in Drosophila S2 cells. (n = 3, mean + SD, **P < 0.01) (K–N) The Sd binding motif is required for Yki-induced target gene expression in vivo. Compared with the controls (K and M), ectopic expression of YkiS168A along the A-P compartment boundary results in dramatic up-regulation of E2–LacZ (L), but not E3–LacZ (N). (O and P) Drosophila S2 cells transfected with Sd and Yki–HA were used for quantification of ChIP-PCRs. (n = 3, mean + SD).