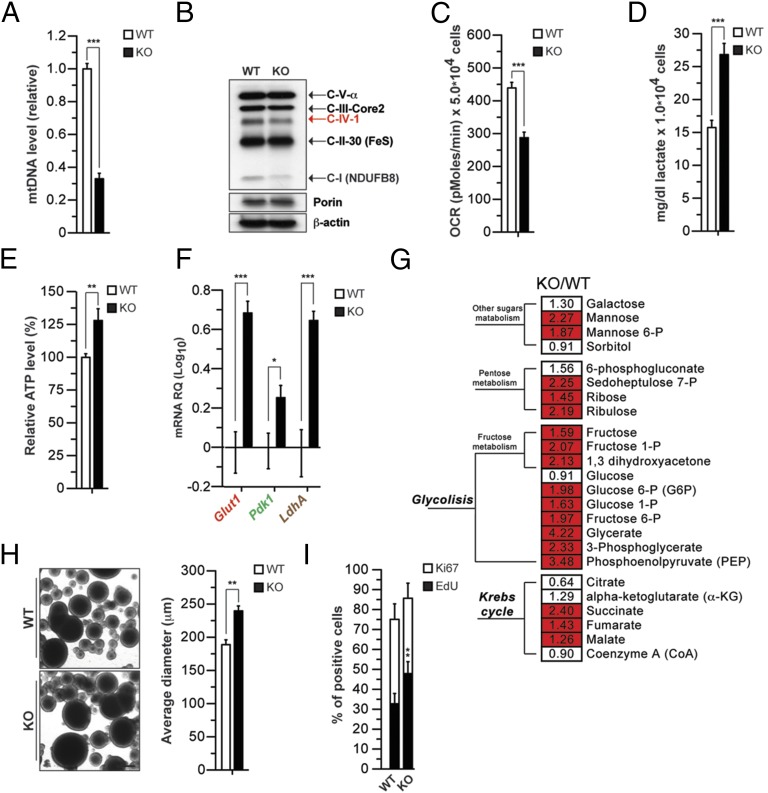
Fig. 2.
Reduction in mitochondrial copy number leads to ETC defects, glycolysis induction, and growth advantage. (A) Relative quantification of mtDNA levels in wild-type (WT) and TK2 KO (KO) NPCs using quantitative RT-PCR (QPCR) (n = 3; ***P < 0.001). (B) Levels of ETC mtDNA-encoded (red) and nuclear-encoded (black) proteins at steady state using an antibody mix against OXPHOS complexes. Porin and β-actin are shown as loading controls for mitochondria and total protein extracts, respectively. (C) OCR in NPCs under basal conditions. Data are average of three independent experiments as mean ± SEM. NPCs were obtained from n = 3 animals for each genotype; ***P < 0.001; lactate (D) and relative ATP levels (E) in NPCs. Measurements were made in triplicate (data are represented as mean ± SEM for n = 3; **P < 0.01, ***P < 0.001). (F) Expression of the genes Glut1, Pdk1, and LdhA in WT and TK2 KO NPCs (expressed as levels over WT cells). (G) Heat map showing the ratio of the metabolite levels between KO and WT NPCs, and their statistical significance of the difference (Welch’s two-samples t test). Cells shaded in red indicate higher metabolite levels in KO NPCs with P < 0.001. Cells not shaded indicate no significant difference (P > 0.05). The number in each cell indicates fold changes over WT. (H) Proliferation of NPCs measured using the neurosphere assay. Neurosphere diameter is increased in restimulated KO NPCs (n = 3; **P < 0.001, Student’s t test). (Scale bar: 100 µm.) (I) Proliferation of NPCs by EdU labeling (2 h) and immunodetection of Ki67. Graph shows quantification of EdU and Ki67.