ABSTRACT
Owing to their small size and enhanced stability, nanobodies derived from camelids have previously been used for the construction of intracellular “nanotraps,” which enable redirection and manipulation of green fluorescent protein (GFP)-tagged targets within living plant and animal cells. By taking advantage of intracellular compartmentalization in the magnetic bacterium Magnetospirillum gryphiswaldense, we demonstrate that proteins and even entire organelles can be retargeted also within prokaryotic cells by versatile nanotrap technology. Expression of multivalent GFP-binding nanobodies on magnetosomes ectopically recruited the chemotaxis protein CheW1-GFP from polar chemoreceptor clusters to the midcell, resulting in a gradual knockdown of aerotaxis. Conversely, entire magnetosome chains could be redirected from the midcell and tethered to one of the cell poles. Similar approaches could potentially be used for building synthetic cellular structures and targeted protein knockdowns in other bacteria.
Importance Intrabodies are commonly used in eukaryotic systems for intracellular analysis and manipulation of proteins within distinct subcellular compartments. In particular, so-called nanobodies have great potential for synthetic biology approaches because they can be expressed easily in heterologous hosts and actively interact with intracellular targets, for instance, by the construction of intracellular “nanotraps” in living animal and plant cells. Although prokaryotic cells also exhibit a considerable degree of intracellular organization, there are few tools available equivalent to the well-established methods used in eukaryotes. Here, we demonstrate the ectopic retargeting and depletion of polar membrane proteins and entire organelles to distinct compartments in a magnetotactic bacterium, resulting in a gradual knockdown of magneto-aerotaxis. This intracellular nanotrap approach has the potential to be applied in other bacteria for building synthetic cellular structures, manipulating protein function, and creating gradual targeted knockdowns. Our findings provide a proof of principle for the universal use of fluorescently tagged proteins as targets for nanotraps to fulfill these tasks.
Importance
Intrabodies are commonly used in eukaryotic systems for intracellular analysis and manipulation of proteins within distinct subcellular compartments. In particular, so-called nanobodies have great potential for synthetic biology approaches because they can be expressed easily in heterologous hosts and actively interact with intracellular targets, for instance, by the construction of intracellular “nanotraps” in living animal and plant cells. Although prokaryotic cells also exhibit a considerable degree of intracellular organization, there are few tools available equivalent to the well-established methods used in eukaryotes. Here, we demonstrate the ectopic retargeting and depletion of polar membrane proteins and entire organelles to distinct compartments in a magnetotactic bacterium, resulting in a gradual knockdown of magneto-aerotaxis. This intracellular nanotrap approach has the potential to be applied in other bacteria for building synthetic cellular structures, manipulating protein function, and creating gradual targeted knockdowns. Our findings provide a proof of principle for the universal use of fluorescently tagged proteins as targets for nanotraps to fulfill these tasks.
INTRODUCTION
Intrabodies are functional fragments derived from full-length antibodies that can be expressed in heterologous hosts and which specifically recognize their antigen within cells. In various eukaryotic systems, they have been demonstrated to be powerful tools that enable the intracellular analysis and manipulation of protein functions (1–5). Among the various types of intrabodies, so-called nanobodies have proven to be particularly useful due to their small size, solubility, enhanced stability, and the relative ease of screening, cloning, and expression procedures (6–9). Nanobodies are routinely derived from camelid heavy-chain antibodies, which lack the light chains present in conventional IgG antibodies and recognize their target by interaction with single VHH (variable domain of heavy chain antibodies) domains (10). After the genetic repertoire of B cells is extracted from an immunized camelid, antigen-binding VHHs can be selected and expressed as highly robust single-domain antibodies. Because of their special topology, nanobodies preferentially bind to concave surfaces of antigens which are often inaccessible to conventional antibodies (11). Examples for nanobody-based applications in living plant and animal cells include the inhibition of enzyme activity through specific binding to the active site (7, 12), modulation of spectral properties of fluorescent proteins (13), and the construction of nanobody-mediated synthetic regulatory circuits (14). Furthermore, different strategies for nanobody-based protein knockdowns have been reported, either by targeting nanobody-bound proteins to degradation pathways (15) or by artificially retargeting interaction partners to specific intracellular localizations (16–18). Artificial relocalization of targeted proteins was either caused by trapping of nanobody-bound proteins in the cytoplasm due to interference with protein translocation to cellular compartments (18) or by specifically anchoring the nanobody to distinct structures and compartments of the eukaryotic cell, such as distinct DNA regions, plasma membranes, or the centrioles of animal cells, resulting in ectopic recruitment of green fluorescent protein (GFP)-tagged targets (16, 17, 19). The application of anchored nanobodies against GFP (GFP-binding protein [GBP]) as an intracellular nanotrap is a particularly versatile tool because of the widespread use of derivatives of this fluorescent tag.
It has been realized only rather recently that prokaryotic cells also contain highly organized subcellular structures (20). Bacteria possess, for example, structural homologs to eukaryotic cytoskeletal elements that define cell shape, structure, and function (21, 22). In addition, they form large supramolecular protein complexes, contain microcompartments, and even biosynthesize canonical membrane-enveloped organelles that show distinct subcellular localization patterns (22–24).
The ability to target proteins intracellularly and possibly even redirect macromolecular complexes to defined subcellular locations in bacteria has great potential for synthetic intracellular scaffolding and targeting of proteins or protein complexes (25, 26). For instance, such techniques could be used for protein knockdowns through spatial separation of interaction partners. Other possible applications are specific targeting of proteins to bacterial subcellular compartments (27–29), the setup of synthetic intracellular gradients (30), or even artificially compartmentalizing and distributing different cellular processes and organelles to distinct subcellular localizations. However, so far there are few tools available that are equivalent to the well-established methods used in eukaryotic cells and that would efficiently fulfill these tasks in bacteria.
One of the most intricate examples of natural compartmentalization in prokaryotic cells is magnetosomes, which are nano-sized ferromagnetic crystals synthesized within intracellular membrane vesicles by magnetotactic bacteria such as Magnetospirillum gryphiswaldense. These organelles are attached to a cytoskeletal filament formed by the actin-like protein MamK and arranged in a chain that is positioned at the midcell (31, 32). The resulting magnetic dipole moment rotates the bacterial cell into alignment with the geomagnetic field, thereby enhancing the movement of the bacteria toward growth-favoring oxygen levels (33). Recently, our lab demonstrated the display of nanobodies on magnetosomes that were functional in recognizing their antigen not only in vitro but also in vivo. Expression of MamC–red fluorescent protein (RFP)-binding protein (RBP) fusions resulted in the recruitment of cytoplasmic RFP to the magnetosomes (34), showing that intracellular localization of soluble heterologous proteins can be manipulated in bacteria. This motivated us to further investigate whether magnetosome anchors can also be used to trap proteins with distinct functions from other cellular compartments. For this purpose, we chose the chemotaxis protein CheW, which is part of the chemoreceptor clusters that are universally found in chemotactic bacteria and typically display a distinct polar localization (35). We demonstrate that CheW1 fused to enhanced GFP (EGFP) can be depleted from cell poles by expression of multivalent GBP nanobodies fused to the magnetosome protein MamC on endogenous levels, resulting in ectopic recruitment of CheW1 to the magnetosome chain of M. gryphiswaldense. Depletion of CheW1 from polar clusters resulted in a gradual impairment of aerotaxis. Intriguingly, the interaction between the magnetosome anchor and polar CheW1-EGFP also led to artificial repositioning of the entire magnetosome chain from its midcell position toward one of the cell poles, indicating that entire organelles can be redirected by nanobodies and tethered to ectopic positions. Our study establishes the application of nanotrap technology for artificial targeting of proteins and even entire organelles to bacterial cells. Similar approaches could be used for building tailored subcellular structures in synthetic biology and for gradual protein knockdowns in other prokaryotic systems.
RESULTS
Recruitment of CheW1-EGFP to magnetosomes with a GBP nanotrap.
In M. gryphiswaldense, CheW1 is encoded within the cheOp1 chemotaxis operon, which was recently demonstrated to control magneto-aerotactic swimming polarity (36). It is well established in various bacteria that CheW acts as a linker and interacts both with the chemoreceptor and the histidine kinase CheA proteins, thereby enhancing the polar chemoreceptor clustering and function (37). First, we replaced the native cheW1 gene with cheW1-egfp via chromosomal insertion. Similarly as observed in other bacteria (38–40), spot-like fluorescent signals originating from EGFP-tagged CheW1 were exclusively found at both cell poles in the wild-type background in fluorescence micrographs (Fig. 1ai and f). This is consistent with previous results of cryoelectron microscopy of M. gryphiswaldense cells, where chemoreceptor complexes were identified near the poles (31). Only in elongated cells close to completion of cell division, two new clusters were formed at the midcell (see Fig. S1 in the supplemental material). In mutant backgrounds either forming magnetosome clusters instead of chains (ΔmamJ mutant) (32) or entirely lacking any magnetite particles (ΔmamM mutant) (41), the same CheW1-EGFP fluorescence localization pattern as in the wild-type background was observed (Fig. 1di; see also Fig. S2b), indicating that polar chemoreceptor localization was independent of the presence and configuration of magnetosome chains, as expected.
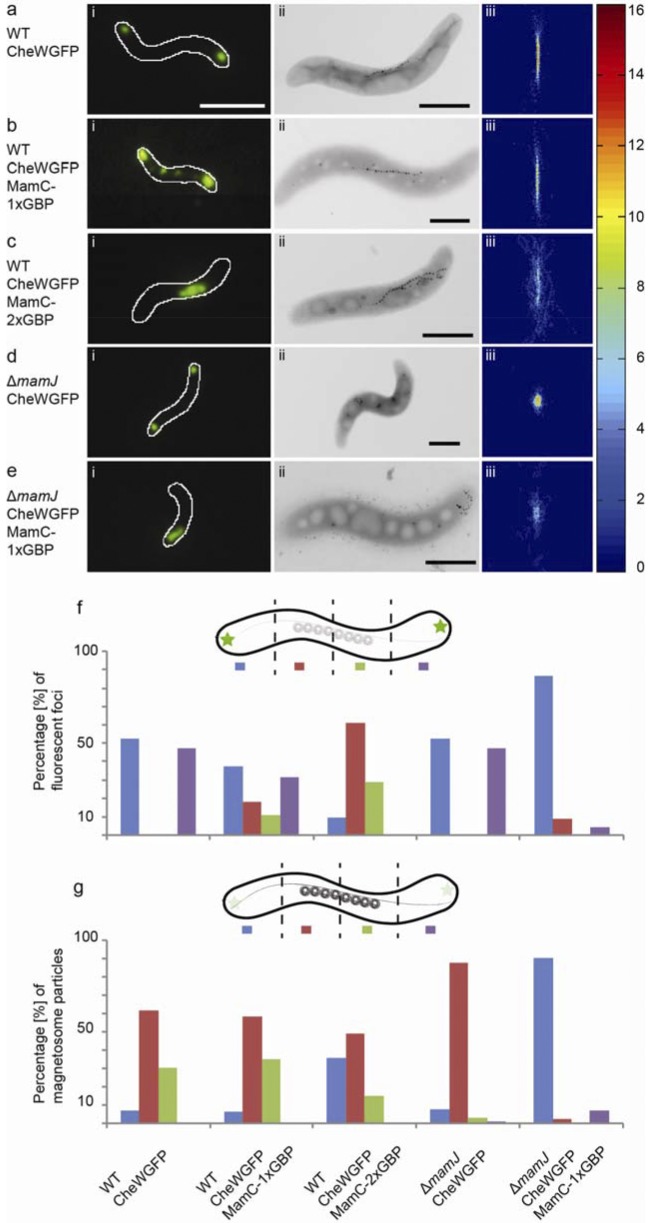
FIG 1 .
Analysis of subcellular CheW1-EGFP and magnetosome localization. Fluorescence (i) and TEM (ii) micrographs of representative M. gryphiswaldense CheW1-EGFP (a), CheW1-EGFP MamC-1×GBP (b), CheW1-EGFP MamC-2×GBP (c), ΔmamJ CheW1-EGFP (d), and ΔmamJ CheW1-EGFP MamC-1×GBP (e) cells. Cells were analyzed by CHAP (iii) and scored for the distribution of fluorescence signal, represented by the percentage of fluorescent foci detected within 4 equidistant compartments (f), and magnetosomes, represented by the percentage of magnetosomes detected within 4 equidistant compartments (g). White scale bar, 2 µm; black scale bar, 1 µm. Twenty cells were aligned by CHAP for each strain, and heat maps display the numbers of magnetosomes. Single cells were segmented into four compartments, and for each strain 20 cells were scored to obtain fluorescence and magnetosome distributions.
Next, we asked whether the localization of CheW1-EGFP was affected by coexpression of a GBP nanobody that had been identified by Rothbauer and colleagues before (42). To trap CheW1-EGFP, GBP was expressed either alone in the cytoplasm (MagGBPcyt) or fused to the abundant magnetosome membrane protein MamC (43), which has routinely been used as a magnetosome anchor for immobilization of various functional moieties, such as EGFP, enzymes, or an RBP (34, 43–46). In addition to the native gbp gene, we used a synthetic allele that was specifically optimized for the expression in M. gryphiswaldense (Magnetospirillum-optimized green-binding protein [maggbp]). MamC was fused to either one single copy of GBP connected to mCherry (mCherry-GBP, also referred to as “chromobody”) (42), and the resulting MamC-mCherry-GBP fusion is referred to as MamC-1×GBP here, or to a tandem copy of maggbp-gbp (resulting in MamC-MagGBP-GBP, referred to as MamC-2×GBP here). All different gbp constructs were inserted into chromosomes of parent strains coexpressing CheW1-EGFP. Western blot analysis of cell extracts of all strains carrying the generated fusions revealed reacting protein bands with expected sizes, indicating that the mono- and bivalent GBP nanobodies were stably expressed on magnetosomes (see Fig. S3 in the supplemental material).
Cytoplasmic expression of unfused MagGBPcyt alone had no effect on the localization of CheW1-EGFP fluorescence in the wild-type background (see Fig. S2d). However, upon coexpression of MamC-1×GBP and CheW1-EGFP, we detected weaker, secondary fluorescent foci at approximately the midcell position in addition to the two polar CheW1-EGFP signals (Fig. 1bi). We scored the number of fluorescent foci in four equidistant sectors along lengths of a representative set of cells and calculated the relative abundance of fluorescence intensity in each of the sectors (see Materials and Methods for details). In contrast to the wild-type background, which displayed only polar foci, about 30% of fluorescence intensity was detected within the cytoplasm upon coexpression of MamC-1×GBP (a representative cell is shown in Fig. 1bi and f). Recruitment of CheW1-EGFP was likely due to interaction with GBP expressed on magnetosomes, as green (CheW1-EGFP) and red (mCherry-tagged magnetosomes) fluorescence signals coincided in all analyzed cells, indicating that direct GBP-EGFP interaction caused the observed redirection of CheW1 (see Fig. S2e and f). In cells coexpressing two GBP copies in tandem (MamC-2×GBP), a single large, nonpolar fluorescence signal was detected in the vast majority of cells. Ninety percent of the CheW1-EGFP fluorescence intensity was shifted toward the midcell (Fig. 1ci and 4), while only 10% of the fluorescence signal remained at the cell pole (Fig. 1f). Instead of the spot-like, exclusively polar foci of the parent strain, a linear fluorescence signal was present near the midcell in most MamC-2×GBP-expressing cells, demonstrating efficient redirection of membrane complex-associated GFP-tagged proteins (Fig. 1ci; see also Fig. S4).
Next, we investigated whether the absence of magnetic nanoparticles would affect the recruitment of CheW1-EGFP through MamC-GBP fusions by analyzing nonmagnetic cells. Due to loss of the magnetosomal iron transporter MamM, ΔmamM cells lack any magnetite crystals but still produce empty magnetosome membrane vesicles (41). CheW1-EGFP fluorescence was shifted toward the midcell in the ΔmamM strain coexpressing MamC-2×GBP to the same extent as in the magnetite-containing strains (see Fig. S2c). To analyze whether the configuration of magnetosome chains had an effect on CheW1-EGFP recruitment, we also expressed MamC-1×GBP in the ΔmamJ background, in which the physical interaction of magnetosome chains with the actin-like MamK filaments is abolished (32), resulting in agglomerated clusters rather than linear, well-ordered chains of magnetosomes (Fig. 1dii and eii). In the vast majority of analyzed ΔmamJ MamC-1×GBP cells, the major proportion of CheW1-EGFP fluorescence (>85% of all foci) was located at only 1 cell pole (Fig. 1f) and appeared to be slightly distorted longitudinally (Fig. 1ei).
Effect of CheW1-EGFP recruitment on magnetosome localization.
We noticed that all strains which showed strong CheW1 mislocalization were increasingly affected in their magnetic alignment, as indicated by reduced magnetic response (Cmag) values (e.g., MamC-2×GBP, 0.60 ± 0.07; wild type, 1.24 ± 0.20). The Cmag value provides an optical measure of the relative alignment of cells in a cuvette by applying a strong magnetic field either parallel or perpendicular to the light beam of a photometer. Transmission electron microscopy (TEM) analysis revealed that wild-type cells expressing CheW1-EGFP alone displayed the same magnetosome localization pattern as that of their parent strain (Fig. 1aii). Both automated image analysis by the Chain Analysis Program (CHAP) (47) and manual scoring of magnetosome position (see Materials and Methods for details; Fig. 1g) indicated that the linear chains of magnetosomes were consistently positioned at the midcell and displayed the same configuration as typically observed for the M. gryphiswaldense parent strain (48, 49), with approximately 35 particles per cell that had an average crystal size of 35 to 47 nm (49). Additional cytoplasmic expression of MagGBPcyt in the same background did not affect magnetosome chain configuration (see Fig. S5b). Coexpression of MamC-1×GBP and CheW1-EGFP did not affect the midcell position of magnetosome chains either, but chains were less compact, i.e., particles were more widely spaced, as indicated by the fuzzier, slightly stretched appearance of magnetosome chains in CHAP analysis heat maps (Fig. 1biii). TEM analysis of ΔmamJ cells expressing CheW1-EGFP alone revealed the same magnetosome localization pattern as that of their parent strain (Fig. 1dii). Consistent with the observed shift of the CheW1-EGFP fluorescence toward one pole in the ΔmamJ MamC-1×GBP strain, 90% of magnetosome clusters detected in TEM micrographs were localized at a single cell pole only, while clusters were no longer observed at the midcell or along the cell length, as commonly found in the ΔmamJ parent strain (32, 50) (Fig. 1g). Moreover, the loose magnetosome assemblies observed at the poles were slightly elongated compared to the compact, rounded magnetosome clusters of the parent strain (Fig. 1diii and eiii). This indicated that targeted recruitment and partial rearrangement of magnetosomes were facilitated in cells in which magnetosome particles were no longer bound to the MamK filament by their molecular connector MamJ (33). As observed for mislocalization of CheW1-EGFP fluorescence, in wild-type cells coexpressing divalent tandem fusions of GBP (MamC-2×GBP), magnetosome chains were predominantly drawn to one of the cell poles (Fig. 1cii; see also Fig. S6). Magnetosome chains were even less compact than in the presence of the monovalent nanobody, as reflected by the rather scattered pattern of poorly aligned magnetosome chains (Fig. 1ciii). Consistent with the overall shift of the chain, the mean fraction of magnetosome particles located at one of the cell poles increased from 7 to 36% (Fig. 1g).
Effect of CheW1-EGFP recruitment on chemotaxis of M. gryphiswaldense.
The observed mislocalization of chains also affected the magnetic alignment of swimming cells. While wild-type cells expressing only CheW1-EGFP predominantly swam aligned to the ambient magnetic field, as did the parent strain, a large fraction of cells coexpressing MamC-2×GBP displayed trajectories that were oriented at random angles to the ambient magnetic field (Fig. 2a). As indicated by video microscopy, motility and mean swimming speed were not affected in any of the analyzed strains. Compared to the ΔcheW1 and ΔcheOp1 control strains, in which aerotaxis was entirely abolished, as indicated by the formation of small aerotactic halos in swim plate assays (see Fig. S7) (36), coexpression of cytoplasmic MagGBPcyt and CheW1-EGFP in the wild-type background did not affect the size of swim halos that were virtually identical to those of the parent strain (Fig. 2b and c).
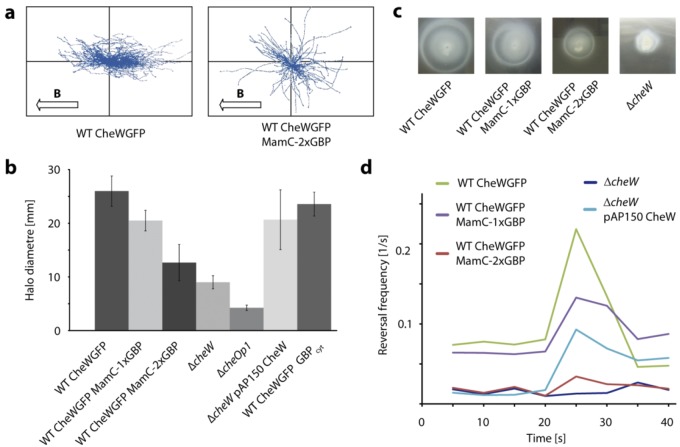
FIG 2 .
Magneto-aerotactic swimming behavior of M. gryphiswaldense strains expressing CheW1-EGFP and MamC-GBP fusions. (a) Magnetic alignment of swimming cells expressing CheW1-EGFP alone or in combination with MamC-2×GBP. A plot of all tracks from a representative video record is shown for each strain. Cells swimming in the gas perfusion chamber were exposed to a homogenous vertical magnetic field of 0.26 mT (B). (b) Average halo diameter of strains expressing MamC-GBP fusions in swim plates (means ± standard deviations [SD] from at least 3 independent replicates). The ΔcheW1 and ΔcheOp1 chemotaxis gene deletion mutants were used as controls. Transcomplementation of the ΔcheW1 mutant strain by constitutive expression of CheW1-EGFP from a plasmid restored chemotactic efficiency to 80% of the wild-type cells expressing CheW1-EGFP at wild-type levels. (c) Halo formation of wild-type CheW1-EGFP, CheW1-EGFP MamC-1×GBP, CheW1-EGFP MamC-2×GBP, and ΔcheW cells in 0.2 % motility agar 3 days after inoculation. (d) Aerotactic reversal response upon abrupt shift from 0% to 2% oxygen in a microscopic gas perfusion chamber. Video records were analyzed by automated tracking software (WimTaxis; Wimasis) to obtain swim tracks and reversal events of individual cells (72), and reversal rates were calculated for 5-s intervals by averaging single-cell data from at least 3 independent recordings.
While the ΔcheW1 strain did not show any response when shifted from anaerobic conditions to 2% oxygen in a microscopic gas perfusion chamber (Fig. 2d) and displayed a straight-swimming phenotype comparable to that of the ΔcheOp1 deletion strain, wild-type cells expressing CheW1-EGFP showed a reaction very similar to that observed in the parent strain (36). The reversal frequency instantaneously rose from less than 0.1 s−1 to more than 0.2 s−1 after microoxic upshift. This was followed by a rapid drop in reversal frequency below prestimulus levels within 15 s (Fig. 2d). However, coexpression of MamC-1×GBP and CheW1-EGFP led to slightly reduced halo sizes in swim plates and a lower number of reversals in response to the oxygen shift. The maximum reversal frequency remained below 0.15 s−1 and peaked at approximately 60% of the wild-type rate. Interestingly, coexpression of MamC-1×GBP also caused delayed adaptation after the shift, as the reversal frequency remained above prestimulus levels within 20 s postshift.
Coexpression of MamC-2×GBP and CheW1-EGFP, which completely depleted CheW1-EGFP from the cell poles as suggested by fluorescence microscopy (Fig. 1ci), also had a dramatic effect on the cells’ switching behavior under anoxic equilibrium conditions and the response elicited by oxygen exposure. The prestimulus reversal frequency was comparable to that of the ΔcheW1 strain and rose only minimally after oxygen upshift to 2% O2, remaining on a very low level (maximum frequency below 0.05 s−1). In conclusion, an increase in the copy number of GBP led to gradually stronger impairment of aerotaxis, eventually reducing the number of reversals in a strain coexpressing CheW1-EGFP and the divalent MamC-2×GBP fusion to the level of a ΔcheW1 null mutant.
DISCUSSION
We investigated the interaction between components of the universal bacterial chemotaxis signaling pathway and nanobodies expressed on the magnetosome organelles of M. gryphiswaldense, which enabled us to easily follow the structural and behavioral effects of artificial recruitment by TEM and fluorescence microscopy (FM) imaging and video microscopy at the single cell level. We observed that by anchoring GBP to the magnetosome membrane, the localization of CheW1-GFP was shifted from the poles to the midcell, i.e., to the typical position of the magnetosome chain. There are two possible explanations for the observed redirection of CheW1 from the polar clusters to the magnetosomes: (i) unbound CheW1, in equilibrium with the receptor-bound form, could be recruited from a cytoplasmic pool, whereas (ii) membrane-bound CheW1 could be directly withdrawn from preexisting polar clusters. CheW is a soluble protein that lacks transmembrane domains but in vitro forms ultrastable ternary complexes together with CheA and chemoreceptors (51). However, in living cells, signaling complexes are weakly dynamic and display slow turnover (of approximately 12 min), as indicated by fluorescence recovery after photobleaching (FRAP) experiments on CheA and CheW constructs (52). Consistent with these observations, it has been suggested that small amounts of CheA might be permanently present in an unbound state in the cytoplasm (53). However, the relative copy numbers of all cluster components are tightly regulated, and since overexpression of CheW leads to impaired chemotactic signaling (due to competitive inhibition of CheA binding to the chemoreceptors) (51), the pool of free CheW in the cytoplasm must be rather small. Thus, it seems most probable that soluble CheW present at low concentrations in the cytoplasm is sequestered by magnetosome-anchored GBP, and over time also those molecules initially bound to the chemoreceptor clusters might gradually be released and trapped at ectopic positions by a strong interaction with the nanobody.
The localization of CheW1-GFP was unaffected by coexpression of cytoplasmic (unfused) GBP in the wild-type background but shifted toward the midcell upon MamC-2×GBP expression in the nonmagnetic ΔmamM strain, which lacks any electron-dense magnetic crystals but still forms empty vesicles of the magnetosome membrane (41). This demonstrates that GBP immobilized on magnetite-free membrane vesicles is sufficiently effective to specifically redirect localized proteins. Furthermore, this suggests that recruitment and retargeting could be achieved in other bacteria lacking magnetosomes by using different spatial determinants as intracellular traps.
Although the presence of magnetic particles was no absolute prerequisite for efficient recruitment, redistribution of CheW was strongly affected by magnetosome chain configuration. Magnetosome clusters were drawn to only one pole in the ΔmamJ background upon expression of MamC-1×GBP and acted as efficient nanotraps for CheW1-GFP. In contrast to the undefined midcell fluorescence caused by partial depletion from polar clusters observed upon expression of MamC-1×GBP in the wild-type background, virtually no CheW1-GFP signal was detected at the midcell or the opposite pole in the ΔmamJ mutant strain. This either might be due to increased avidity of nanobodies concentrated in the tightly clustered magnetosome assemblies or might reflect a stochastic shift of CheW diffusion equilibrium due to the concentration of two sinks (i.e., the native chemoreceptor cluster and the artificial magnetosomal nanobody cluster) at a single pole (Fig. 3, bottom right).
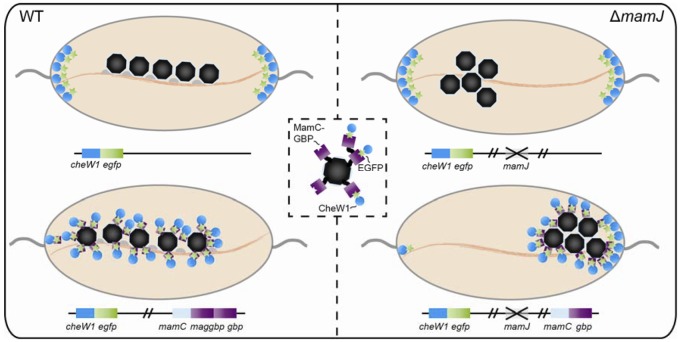
FIG 3 .
Model of MamC-GBP and CheW1-EGFP interaction. CheW1-EGFP localizes distinctly at the cell poles if expressed chromosomally in the wild-type and ΔmamJ backgrounds (top right and left). If MamC-GBP is coexpressed in the wild type (bottom left), CheW1-EGFP is depleted completely from the poles. Expression of MamC-GBP in the ΔmamJ CheW1-EGFP background leads to recruitment of whole magnetosomes to the cell poles (bottom right). Expression of monovalent and divalent nanobodies on a magnetosomes and interaction with CheW1-EGFP is illustrated in the inset. Expressed proteins are illustrated in same colors as genes.
Interaction of MamC-GBP and CheW1-GFP reciprocally affected configuration and positioning of the magnetosome chain. Binding of CheW1-GFP to magnetosome particles disturbed their proper alignment into regular, densely spaced chains. Increasing the expression of GBP (MamC-1×GBP and -2×GBP) also gradually increased the interparticle spacing, possibly by additional protein bound to the magnetosome surface which might weaken the magnetostatic interactions between particles. Overexpression of MamC-2×GBP in the wild-type background caused a nearly complete shift of the magnetosome chains toward the poles, with the majority of magnetosome chains originating at polar or subpolar positions (see Fig. S6), which was probably caused by redirecting and tethering the chains to a fraction of membrane-bound CheW1-GFP remaining at the cell pole (Fig. 3, bottom left). Although the magnetosome chain of wild-type cells generally occupies the midcell position, it becomes mobilized during cell division, when the chain is split and repositioned by MamK dynamics to the midcell of daughter cells (50). We found that magnetosome chain localization was most severely impaired in cells lacking the acidic MamJ protein, which is assumed to connect magnetosome particles to the cytoskeletal magnetosome filament formed by the actin-like MamK protein (32). In the ΔmamJ background, overexpression of the monovalent nanobody was already sufficient to rearrange (Fig. 1eiii) and recruit (Fig. 1eii) the magnetosome cluster to 1 cell pole (Fig. 3, bottom right). The increased intracellular mobility of ΔmamJ magnetosome clusters might be explained by a lack of the presumed MamK-mediated interactions with divisome constituents (50). In wild-type cells, these interactions need to be overcome by interaction with polar CheW, whereas in ΔmamJ cells, magnetosome redirection is facilitated because MamK-magnetosome interactions are abolished.
The level of CheW1-GFP recruitment clearly depended on gene dosage. While redirection of CheW1-GFP was only partial in cells expressing MamC-1×GBP, overexpression of MamC-2×GBP caused a complete shift of CheW1-GFP localization toward the midcell. There is precedence for significantly increased avidity (500×) of a nanobody consisting of a fusion of two identical domains compared to the monovalent nanobody (54). Similarly, in our experiments, the binding of CheW1-GFP to the monovalent GBP was apparently comparable to the in vivo turnover of the chemoreceptor-CheW complexes, since polar- and midcell-localized CheW1-GFP could be detected. In contrast, the avidity of the bivalent nanobody was much stronger, more endogenous CheW1-GFP was bound, and the equilibrium was shifted toward the GBP-bound state.
In addition to demonstrating efficient redirection of entire organelles to distinct locations, we observed that ectopic redirection of CheW1-GFP also gradually modulated chemotactic efficiency of M. gryphiswaldense cells. While chemoreceptors readily form complexes in the absence of CheA and CheW, the latter is essential for stabilizing native CheA-receptor interactions and lattice formation (37, 53). Partial depletion of CheW1 gradually reduced chemotactic efficiency, while expression of the bivalent nanobody essentially phenocopied the deletion of cheW1 (Fig. 2c). As GBP expressed in the cytoplasm had no effect on aerotaxis, this was not due to inactivation of CheW1-GFP but caused by redirection and depletion from its native polar environment. Although bacteria do not display the same level of compartmentalization as eukaryotic cells, the functionality of many bacterial proteins similarly depends on their localization. Our results show that testing protein function by manipulating its subcellular localization as applied to eukaryotic systems (2, 18) can be extended to much smaller bacterial cells and be used to efficiently modulate protein function by subcellular retargeting.
Compared to other approaches for silencing or manipulating the expression of selected genes at the DNA or RNA (55, 56) level, the biggest advantage of regulating gene expression at the protein level is that there is no change of mRNA transcript or native protein expression level (57). Especially for bacterial genes encoded in operons, gradual knockdown of individual proteins might be difficult to achieve at the transcriptional level, if polar effects on transcription of downstream genes are to be avoided. Additionally, it would be desirable to develop inducible systems, e.g., to gradually control in vivo the stoichiometry of proteins in larger clusters. This might facilitate the study of complex regulatory pathways, such as cell division or cell differentiation processes in other bacteria.
Intrabodies are well established as powerful tools in eukaryotic cells for trapping soluble proteins at defined subcellular locations (16–18) or for inhibition of protein function (12). Although recombinant nanobodies can be produced easily in bacteria such as Escherichia coli (10), to date the use of intrabodies in bacterial cells has been restricted to only very few studies. Two early publications reported the intracellular expression of single-chain Fv antibody fragments (e.g., to block transcriptional activation) (58, 59), and more recently nanobodies have been applied in bacteria to inhibit enzyme activity (60). However, in these approaches, intrabodies were not anchored to defined positions, and inhibition of enzymes was achieved by neutralization rather than redirection to completely different compartments of the cell. Although for proof of principle we took advantage of the specific compartmentalization in M. gryphiswaldense, in which the magnetosomes provide a natural anchor for setting up an intracellular nanotrap, this approach could also be extended and adapted for application in other bacteria. By using universal tags like GFP for recruitment, many proteins can be targeted with the same nanobody by applying the same strategy, obviating the need of camelid immunization and screening of whole libraries. Multiple other applications are possible, because GFP fusion proteins can be combined with any cellular anchor point, such as subcellular locations (e.g., poles, midcell), specific protein complexes, compartments, organelles, or other spatial determinants. For instance, potential applications of our approach in bacteria could be building synthetic cellular structures (e.g., artificial tethering of heterologously expressed bacterial microcompartments) or compartmentalization of biosynthetic pathways, which can dramatically increase production by restricting reactions spatially to subcellular compartments (61, 62).
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table S1 and S2 in the supplemental material. M. gryphiswaldense strains were grown microaerobically with 1% oxygen in modified flask standard medium (FSM) at 30°C (63) and moderate shaking (120 rpm). E. coli strains were cultivated as previously described (64). For growth of E. coli WM3064 (W. Metcalf, unpublished data) or BW29427 (K. Datsenko and B. L. Wanner, unpublished data), 1 mM dl-α,ε-diaminopimelic acid (DAP) was added to lysogeny broth medium (LB). Strains were routinely cultured on plates solidified with 1.5% (wt/vol) agar. For strains carrying recombinant plasmids, media were supplemented with 25 µg ⋅ ml−1 kanamycin and 50 µg ⋅ ml−1 ampicillin (Amp) for E. coli strains and 5 µg ⋅ ml−1 kanamycin (km) for M. gryphiswaldense strains.
For the preparation of swim plates, only 0.2% agar was used, the concentration of carbon source (lactate) was lowered to 1.5 mM, and peptone was omitted from FSM. Five microliters of overnight culture was pipetted into the swim agar, and plates were incubated under microoxic conditions for 2 days (protocol modified from that of Schultheiss et al. [65]).
Molecular and genetic techniques.
Oligonucleotides were purchased from Sigma-Aldrich (Steinheim, Germany), and sequences can be supplied on request. Plasmids were constructed by standard recombinant techniques as described in detail below. All constructs were sequenced on an ABI 3730 capillary sequencer (Applied Biosystems, Darmstadt, Germany), utilizing BigDye Terminator v3.1. Sequence data were analyzed with Software Vector NTI Advance 11.5 (Invitrogen, Darmstadt, Germany). The GBP nanobody (42) was provided by ChromoTek GmbH (Planegg-Martinsried), and a synthetic GBP was specifically optimized for the expression in M. gryphiswaldense with respect to its codon usage and purchased from ATG:biosynthetics (Merzhausen, Germany).
Construction of plasmids for chromosomal gene insertion, deletion, and fusion.
For chromosomal exchange of cheW1 against cheW1-egfp, the fluorescence marker was fused via overlap extension PCR (66) to cheW1 and to a 1,000-bp downstream fragment of the gene. The fused product was inserted into pORFM, and the native cheW1 copy was exchanged chromosomally against cheW1-egfp by homologous recombination facilitated by GalK counterselection (67). Deletion of cheW1 was achieved following a similar strategy by fusion of approximately 1,000-bp fragments upstream and downstream of cheW1 connected by 12 nonsense bp replacing the native cheW1. For complementation of cheW1 deletion, cheW1 was amplified from the genome and inserted into pAP150 (46).
All mamC-gbp fusions were chromosomally introduced by transposition; therefore, all gene fusions created by overlap PCR were inserted into transposable pSB6 and pSB7 plasmids (46).
Analytical methods.
Magnetic reaction of cells was validated by light microscopy applying a bar magnet. Optical density (OD) and magnetic response (Cmag) of exponentially growing cells were measured photometrically at 565 nm as previously reported (68). For Cmag measurement, a magnetic field of approximately 70 mT was used.
Biochemical methods.
Polyacrylamide gels were prepared according to the method of Laemmli (69). Strains were grown overnight and spun down via centrifugation, OD565 was set to 10, and 20 µl was loaded onto 12% (wt/vol) SDS gels and analyzed via immunoblotting. Proteins were electroblotted onto polyvinylidene difluoride (PVDF) membranes (Roth, Germany). Membranes were blocked for 1 h at room temperature with blocking solution (2.5% [wt/vol] milk powder in Tris-buffered saline [TBS] [50 mM Tris-HCl, pH 7.6, and 150 mM NaCl]) and incubated for another hour with primary rabbit anti-MamC IgG antibody (1:500 dilution; Santa Cruz, CA). Membranes were washed 4 times with TBS for 5 min and incubated with a secondary alkaline phosphatase-labeled goat anti-rabbit IgG antibody (1:2,000 dilution; Promega, United States) for 45 min. Membranes were washed 4 times with TBS for 5 min, and immunoreactive proteins were visualized with nitroblue tetrazolium (NBT)/5-bromo-4-chloro-3-indolylphosphate (BCIP) (Roche kit).
Phase contrast and fluorescence microscopy.
Strains with genomic CheW1-EGFP fusions and additional MamC-GBP fusions were grown in 1 ml FSM in 24-well plates for 16 h at 30°C and 1% O2 without agitation. For microscopy, cells were immobilized on agarose pads (phosphate-buffered saline [PBS] buffer supplemented with 1% agarose) and imaged with an Olympus BX81 microscope equipped with a 100 UPLSAPO100XO objective (numerical aperture of 1.40) and a Hamamatsu Orca AG camera. The Olympus xcellence pro software was used to capture and analyze images.
To analyze relative positions of fluorescent foci, we manually segmented each cell along its long axis into four equal sectors and scored the fluorescent foci within each sector. The strongest fluorescence signal(s) was scored as “++,” and weaker signals were scored as “+.” Since the orientation of imaged cells was random and in many cases the distribution of fluorescent foci was not perfectly symmetric, we rotated the cells where necessary so that the sectors with the highest cumulated score were sectors 1 and 2. We then calculated relative frequencies of fluorescent focus positions based on the ratio of cumulated scoring points of all analyzed cells per sector divided by the total number of scoring points in all cells.
Transmission electron microscopy.
Magnetosome chain localization was examined by transmission electron microscopy (TEM), for which cells were concentrated via centrifugation and adsorbed onto carbon-coated copper grids. Cells were imaged with an FEI Morgagni 268 (FEI, Eindhoven, Netherlands) electron microscope at an accelerating voltage of 80 kV. For analysis of magnetosome alignment and chain compactness, we used the CHAP script implemented in Matlab and ran the program for 20 cells for each strain (48). For analysis of magnetosome position, we manually segmented each cell along its long axis into four equal sectors and scored the number of magnetosomes within each sector. Since the orientation of imaged cells was random and in many cases the distributions of magnetosomes were not perfectly symmetric, we rotated the cells where necessary so that the sector with the most magnetosomes scored was either sector 1 or 2. We then calculated relative frequencies of magnetosome positions based on the ratio of cumulated magnetosomes of all analyzed cells per sector divided by the total number of magnetosomes in all cells.
Video microscopy and analysis of swimming parameters.
The swimming behavior of cells was analyzed and recorded using dark-field microscopy on an upright Zeiss Axioplan microscope (Zeiss, Jena, Germany) at ×100 magnification. All microscopic motility experiments were performed within a microscopic gas perfusion chamber (Ludin Chamber, Life Imaging Services) that was equilibrated with variably moisturized and precisely adjusted O2-N2 gas mixtures containing between 0 and 2% oxygen (70).
Homogeneous conditions were maintained by using strongly diluted cell suspensions (OD of 0.005) and exposing cell suspensions to a constant gas flow of 50 ml ⋅ min−1, protected against turbulence by placing a gas-permeable agar sheet on top.
Videos were recorded with a UK1158-M camera (EHD, Damme, Germany) at a frame rate of 15 frames per second and a standard resolution of 1,360 by 1,024 pixels using VirtualDub software. Dark-field video records were analyzed by a custom-made automated tracking software (WimTaxis—Bacteria Tracking; Wimasis GmbH, Munich, Germany) specifically adapted to determine the basic swimming characteristics. The software automatically detected swimming reversals and provided the x-y coordinates of every tracked cell for each frame.
The minimum track length was set to be 50 frames. Within the usual tracking times (depending on the time bacteria stayed in the viewing field [usually fewer than 10 s]), reversals generally were too infrequent to simply average the reversal rates for single cells. Therefore, the reversal frequency analysis for each experiment was performed at the population level, and all detected reversals were divided by the total respective tracking time (sum of the temporal length of all tracks) to obtain the population average.
To analyze the cells’ reaction to oxygen shifts, the gas stream was manually switched between oxic and anoxic. For this purpose, we equipped our setup with a three-way valve and a flowmeter to adjust the flow of N2 gas to 50 ml ⋅ min−1 (70). Cells were first equilibrated for 3 min under anoxic conditions before the video recording was started. After 20 s, the gas flow was shifted to 2% O2, and cells were recorded for an additional 20 s. To determine the average reversal frequency over time, the numbers of detected reversals within 5-s intervals were added from three independent video recordings and normalized to the total corresponding tracking time.
SUPPLEMENTAL MATERIAL
CheW1-EGFP localization in a dividing cell. Representative differential interference contrast (DIC) and fluorescence micrographs of a cell chromosomally expressing CheW1-EGFP close to cell division. CheW1-EGFP localizes distinctly at the cell poles and forms two new foci at the midcell at the expected position of septum formation. Scale bar, 2 µm. Download
Schematics (a) and fluorescence micrograph of M. gryphiswaldense ΔmamM mutant cells expressing CheW1-EGFP alone (b) or in combination with MamC-2×GBP (c) and wild-type cells expressing chromosomal CheW1-EGFP and cytoplasmic GBP (d) or CheW1-EGFP and MamC-mCherry-GBP (e and f). Scale bar, 2 µm. Download
Western blot of M. gryphiswaldense cells expressing CheW1-EGFP (1), CheW1-EGFP MamC-mCherry-GBP (55.43 kDa) (2), and CheW1-EGFP MamC-2×GBP (41.07 kDa); protein bands are indicated by black arrowheads (3). The native, unfused MamC (12.35 kDa) is present in all strains, indicated by the white arrowhead. Additional bands are visible for all strains expressing MamC fusions, proving stable expression. MamC was detected using rabbit anti-MamC IgG as the primary antibody and goat anti-rabbit IgG alkaline phosphatase antibodies as the secondary antibody. A PageRuler prestained protein ladder from Fermentas was used as a standard. Download
Fluorescence distribution in fluorescence micrographs of M. gryphiswaldense cells expressing chromosomal CheW1-EGFP MamC-2×GBP. Scale bar, 2 µm. Download
TEM micrographs of the M. gryphiswaldense ΔcheW1 strain (a) and cells expressing CheW1-EGFP and cytoplasmic MagGBP (b) or CheW1-EGFP and MamC-mCherry-GBP (c). Scale bar, 1 µm. Download
TEM micrographs of M. gryphiswaldense cells expressing CheW1-EGFP and MamC-2×GBP. Scale bar, 2 µm. Download
Swim halos of M. gryphiswaldense ΔcheOp1 cells and wild-type cells coexpressing either CheW1-EGFP and MagGBPcyt or CheW1-EGFP and MamC-mCherry-GBP. Download
Plasmids used in this study.
Strains used in this study.
ACKNOWLEDGMENTS
We thank ChromoTek GmbH for providing the GBP nanobody. We also thank M. Eibauer and A. Heins for help with CHAP implementation.
This work was funded by DFG grants Schu1080/9-1, 15-1, and 16-1 to D.S.
S.B., F.P., and D.S. designed the research; S.B., F.P., and J.H. performed the research; S.B., F.P., and D.S. analyzed the data; S.B., F.P., and D.S. wrote the paper.
Footnotes
Citation Borg S, Popp F, Hofmann J, Leonhardt H, Rothbauer U, Schüler D. 2015. An intracellular nanotrap redirects proteins and organelles in live bacteria. mBio 6(1):e02117-14. doi:10.1128/mBio.02117-14.
REFERENCES
- 1.Carlson JR, Weissman IL. 1988. Molecular tools for inactivating a yeast enzyme in vivo. Mol Cell Biol 8:2647–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaiser PD, Maier J, Traenkle B, Emele F, Rothbauer U. 2014. Recent progress in generating intracellular functional antibody fragments to target and trace cellular components in living cells. Biochim Biophys Acta 1844:1933–1942. doi: 10.1016/j.bbapap.2014.04.019. [DOI] [PubMed] [Google Scholar]
- 3.Marasco WA, Haseltine WA, Chen SY. 1993. Design, intracellular expression, and activity of a human anti-human immunodeficiency virus type 1 Gp120 single-chain antibody. Proc Natl Acad Sci U S A 90:7889–7893. doi: 10.1073/pnas.90.16.7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tavladoraki P, Benvenuto E, Trinca S, De Martinis D, Cattaneo A, Galeffi P. 1993. Transgenic plants expressing a functional single-chain Fv antibody are specifically protected from virus attack. Nature 366:469–472. doi: 10.1038/366469a0. [DOI] [PubMed] [Google Scholar]
- 5.Wesolowski J, Alzogaray V, Reyelt J, Unger M, Juarez K, Urrutia M, Cauerhff A, Danquah W, Rissiek B, Scheuplein F, Schwarz N, Adriouch S, Boyer O, Seman M, Licea A, Serreze DV, Goldbaum FA, Haag F, Koch-Nolte F. 2009. Single domain antibodies: promising experimental and therapeutic tools in infection and immunity. Med Microbiol Immunol 198:157–174. doi: 10.1007/s00430-009-0116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodelón G, Palomino C, Fernández LÁ. 2013. Immunoglobulin domains in Escherichia coli and other enterobacteria: from pathogenesis to applications in antibody technologies. FEMS Microbiol Rev 37:204–250. doi: 10.1111/j.1574-6976.2012.00347.x. [DOI] [PubMed] [Google Scholar]
- 7.Lauwereys M, Arbabi Ghahroudi M, Desmyter A, Kinne J, Hölzer W, De Genst E, Wyns L, Muyldermans S. 1998. Potent enzyme inhibitors derived from dromedary heavy-chain antibodies. EMBO J 17:3512–3520. doi: 10.1093/emboj/17.13.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pellis M, Pardon E, Zolghadr K, Rothbauer U, Vincke C, Kinne J, Dierynck I, Hertogs K, Leonhardt H, Messens J, Muyldermans S, Conrath K. 2012. A bacterial-two-hybrid selection system for one-step isolation of intracellularly functional nanobodies. Arch Biochem Biophys 526:114–123. doi: 10.1016/j.abb.2012.04.023. [DOI] [PubMed] [Google Scholar]
- 9.De Meyer T, Muyldermans S, Depicker A. 2014. Nanobody-based products as research and diagnostic tools. Trends Biotechnol 32:263–270. doi: 10.1016/j.tibtech.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Muyldermans S. 2013. Nanobodies: natural single-domain antibodies. Annu Rev Biochem 82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 11.De Genst E, Silence K, Decanniere K, Conrath K, Loris R, Kinne J, Muyldermans S, Wyns L. 2006. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc Natl Acad Sci U S A 103:4586–4591. doi: 10.1073/pnas.0505379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jobling SA, Jarman C, Teh MM, Holmberg N, Blake C, Verhoeyen ME. 2003. Immunomodulation of enzyme function in plants by single-domain antibody fragments. Nat Biotechnol 21:77–80. doi: 10.1038/nbt772. [DOI] [PubMed] [Google Scholar]
- 13.Kirchhofer A, Helma J, Schmidthals K, Frauer C, Cui S, Karcher A, Pellis M, Muyldermans S, Casas-Delucchi CS, Cardoso MC, Leonhardt H, Hopfner KP, Rothbauer U. 2010. Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol 17:133–138. doi: 10.1038/nsmb.1727. [DOI] [PubMed] [Google Scholar]
- 14.Tang JC, Szikra T, Kozorovitskiy Y, Teixiera M, Sabatini BL, Roska B, Cepko CL. 2013. A nanobody-based system using fluorescent proteins as scaffolds for cell-specific gene manipulation. Cell 154:928–939. doi: 10.1016/j.cell.2013.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caussinus E, Kanca O, Affolter M. 2012. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat Struct Mol Biol 19:117–121. doi: 10.1038/nsmb.2180. [DOI] [PubMed] [Google Scholar]
- 16.Herce HD, Deng W, Helma J, Leonhardt H, Cardoso MC. 2013. Visualization and targeted disruption of protein interactions in living cells. Nat Commun 4:2660. doi: 10.1038/ncomms3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. 2008. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics 7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Schornack S, Fuchs R, Huitema E, Rothbauer U, Lipka V, Kamoun S. 2009. Protein mislocalization in plant cells using a GFP-binding chromobody. Plant J 60:744–754. doi: 10.1111/j.1365-313X.2009.03982.x. [DOI] [PubMed] [Google Scholar]
- 19.Spira F, Mueller NS, Beck G, von Olshausen P, Beig J, Wedlich-Söldner R. 2012. Patchwork organization of the yeast plasma membrane into numerous coexisting domains. Nat Cell Biol 14:640–648. doi: 10.1038/ncb2487. [DOI] [PubMed] [Google Scholar]
- 20.Gitai Z. 2005. The new bacterial cell biology: moving parts and subcellular architecture. Cell 120:577–586. doi: 10.1016/j.cell.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 21.Margolin W. 2009. Sculpting the bacterial cell. Curr Biol 19:R812–R822. doi: 10.1016/j.cub.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shapiro L, McAdams HH, Losick R. 2009. Why and how bacteria localize proteins. Science 326:1225–1228. doi: 10.1126/science.1175685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cornejo E, Abreu N, Komeili A. 2014. Compartmentalization and organelle formation in bacteria. Curr Opin Cell Biol 26:132–138. doi: 10.1016/j.ceb.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murat D, Byrne M, Komeili A. 2010. Cell biology of prokaryotic organelles. Cold Spring Harbor Perspect Biol 2:a000422. doi: 10.1101/cshperspect.a000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen AH, Silver PA. 2012. Designing biological compartmentalization. Trends Cell Biol 22:662–670. doi: 10.1016/j.tcb.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Bashor CJ, Horwitz AA, Peisajovich SG, Lim WA. 2010. Rewiring cells: synthetic biology as a tool to interrogate the organizational principles of living systems. Annu Rev Biophys 39:515–537. doi: 10.1146/annurev.biophys.050708.133652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choudhary S, Quin MB, Sanders MA, Johnson ET, Schmidt-Dannert C. 2012. Engineered protein nano-compartments for targeted enzyme localization. PLoS One 7:e33342. doi: 10.1371/journal.pone.0033342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frank S, Lawrence AD, Prentice MB, Warren MJ. 2013. Bacterial microcompartments moving into a synthetic biological world. J Biotechnol 163:273–279. doi: 10.1016/j.jbiotec.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Kerfeld CA, Heinhorst S, Cannon GC. 2010. Bacterial microcompartments. Annu Rev Microbiol 64:391–408. doi: 10.1146/annurev.micro.112408.134211. [DOI] [PubMed] [Google Scholar]
- 30.Chau AH, Walter JM, Gerardin J, Tang C, Lim WA. 2012. Designing synthetic regulatory networks capable of self-organizing cell polarization. Cell 151:320–332. doi: 10.1016/j.cell.2012.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katzmann E, Scheffel A, Gruska M, Plitzko JM, Schüler D. 2010. Loss of the actin-like protein MamK has pleiotropic effects on magnetosome formation and chain assembly in Magnetospirillum gryphiswaldense. Mol Microbiol 77:208–224. doi: 10.1111/j.1365-2958.2010.07202.x. [DOI] [PubMed] [Google Scholar]
- 32.Scheffel A, Gruska M, Faivre D, Linaroudis A, Graumann PL, Plitzko JM, Schüler D. 2006. Corrigendum: an acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature 441:248. doi: 10.1038/nature04777. [DOI] [PubMed] [Google Scholar]
- 33.Faivre D, Schüler D. 2008. Magnetotactic bacteria and magnetosomes. Chem Rev 108:4875–4898. doi: 10.1021/cr078258w. [DOI] [PubMed] [Google Scholar]
- 34.Pollithy A, Romer T, Lang C, Müller FD, Helma J, Leonhardt H, Rothbauer U, Schüler D. 2011. Magnetosome expression of functional camelid antibody fragments (nanobodies) in Magnetospirillum gryphiswaldense. Appl Environ Microbiol 77:6165–6171. doi: 10.1128/AEM.05282-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, Müller A, Iancu CV, Murphy GE, Dobro MJ, Zhulin IB, Jensen GJ. 2009. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci U S A 106:17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popp F, Armitage JP, Schüler D. 2014. Polarity of bacterial magnetotaxis is controlled by aerotaxis through a common sensory pathway. Nat Commun 5:5398. doi: 10.1038/ncomms6398. [DOI] [PubMed] [Google Scholar]
- 37.Kentner D, Thiem S, Hildenbeutel M, Sourjik V. 2006. Determinants of chemoreceptor cluster formation in Escherichia coli. Mol Microbiol 61:407–417. doi: 10.1111/j.1365-2958.2006.05250.x. [DOI] [PubMed] [Google Scholar]
- 38.Maddock JR, Shapiro L. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 39.Wadhams GH, Warren AV, Martin AC, Armitage JP. 2003. Targeting of two signal transduction pathways to different regions of the bacterial cell. Mol Microbiol 50:763–770. doi: 10.1046/j.1365-2958.2003.03716.x. [DOI] [PubMed] [Google Scholar]
- 40.Ringgaard S, Schirner K, Davis BM, Waldor MK. 2011. A family of ParA-like ATPases promotes cell pole maturation by facilitating polar localization of chemotaxis proteins. Genes Dev 25:1544–1555. doi: 10.1101/gad.2061811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uebe R, Junge K, Henn V, Poxleitner G, Katzmann E, Plitzko JM, Zarivach R, Kasama T, Wanner G, Pósfai M, Böttger L, Matzanke B, Schüler D. 2011. The cation diffusion facilitator proteins MamB and MamM of Magnetospirillum gryphiswaldense have distinct and complex functions, and are involved in magnetite biomineralization and magnetosome membrane assembly. Mol Microbiol 82:818–835. doi: 10.1111/j.1365-2958.2011.07863.x. [DOI] [PubMed] [Google Scholar]
- 42.Rothbauer U, Zolghadr K, Tillib S, Nowak D, Schermelleh L, Gahl A, Backmann N, Conrath K, Muyldermans S, Cardoso MC, Leonhardt H. 2006. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat Methods 3:887–889. doi: 10.1038/nmeth953. [DOI] [PubMed] [Google Scholar]
- 43.Lang C, Schüler D. 2008. Expression of green fluorescent protein fused to magnetosome proteins in microaerophilic magnetotactic bacteria. Appl Environ Microbiol 74:4944–4953. doi: 10.1128/AEM.00231-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grünberg K, Müller EC, Otto A, Reszka R, Linder D, Kube M, Reinhardt R, Schüler D. 2004. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl Environ Microbiol 70:1040–1050. doi: 10.1128/AEM.70.2.1040-1050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohuchi S, Schüler D. 2009. In vivo display of a multisubunit enzyme complex on biogenic magnetic nanoparticles. Appl Environ Microbiol 75:7734–7738. doi: 10.1128/AEM.01640-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Borg S, Hofmann J, Pollithy A, Lang C, Schüler D. 2014. New vectors for chromosomal integration enable high-level constitutive or inducible magnetosome expression of fusion proteins in Magnetospirillum gryphiswaldense. Appl Environ Microbiol 80:2609–2616. doi: 10.1128/AEM.00192-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katzmann E, Eibauer M, Lin W, Pan Y, Plitzko JM, Schüler D. 2013. Analysis of magnetosome chains in magnetotactic bacteria by magnetic measurements and automated image analysis of electron micrographs. Appl Environ Microbiol 79:7755–7762. doi: 10.1128/AEM.02143-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jogler C, Schüler D. 2009. Genomics, genetics, and cell biology of magnetosome formation. Annu Rev Microbiol 63:501–521. doi: 10.1146/annurev.micro.62.081307.162908. [DOI] [PubMed] [Google Scholar]
- 49.Lohsse A, Ullrich S, Katzmann E, Borg S, Wanner G, Richter M, Voigt B, Schweder T, Schüler D. 2011. Functional analysis of the magnetosome island in Magnetospirillum gryphiswaldense: the mamAB operon is sufficient for magnetite biomineralization. PLoS One 6:e25561. doi: 10.1371/journal.pone.0025561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katzmann E, Müller FD, Lang C, Messerer M, Winklhofer M, Plitzko JM, Schüler D. 2011. Magnetosome chains are recruited to cellular division sites and split by asymmetric septation. Mol Microbiol 82:1316–1329. doi: 10.1111/j.1365-2958.2011.07874.x. [DOI] [PubMed] [Google Scholar]
- 51.Erbse AH, Falke JJ. 2009. The core signaling proteins of bacterial chemotaxis assemble to form an ultrastable complex. Biochemistry 48:6975–6987. doi: 10.1021/bi900641c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schulmeister S, Ruttorf M, Thiem S, Kentner D, Lebiedz D, Sourjik V. 2008. Protein exchange dynamics at chemoreceptor clusters in Escherichia coli. Proc Natl Acad Sci U S A 105:6403–6408. doi: 10.1073/pnas.0710611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Briegel A, Wong ML, Hodges HL, Oikonomou CM, Piasta KN, Harris MJ, Fowler DJ, Thompson LK, Falke JJ, Kiessling LL, Jensen GJ. 2014. New insights into bacterial chemoreceptor array structure and assembly from electron cryotomography. Biochemistry 53:1575–1585. doi: 10.1021/bi5000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coppieters K, Dreier T, Silence K, de Haard H, Lauwereys M, Casteels P, Beirnaert E, Jonckheere H, Van de Wiele C, Staelens L, Hostens J, Revets H, Remaut E, Elewaut D, Rottiers P. 2006. Formatted anti-tumor necrosis factor α VHH proteins derived from camelids show superior potency and targeting to inflamed joints in a murine model of collagen-induced arthritis. Arthritis Rheum 54:1856–1866. doi: 10.1002/art.21827. [DOI] [PubMed] [Google Scholar]
- 55.Man S, Cheng R, Miao C, Gong Q, Gu Y, Lu X, Han F, Yu W. 2011. Artificial trans-encoded small non-coding RNAs specifically silence the selected gene expression in bacteria. Nucleic Acids Res 39:e50. doi: 10.1093/nar/gkr034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Na D, Yoo SM, Chung H, Park H, Park JH, Lee SY. 2013. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat Biotechnol 31:170–174. doi: 10.1038/nbt.2461. [DOI] [PubMed] [Google Scholar]
- 57.Melchionna T, Cattaneo A. 2007. A protein silencing switch by ligand-induced proteasome-targeting intrabodies. J Mol Biol 374:641–654. doi: 10.1016/j.jmb.2007.09.053. [DOI] [PubMed] [Google Scholar]
- 58.Garcillán-Barcia MP, Jurado P, González-Pérez B, Moncalián G, Fernández LA, de la Cruz F. 2007. Conjugative transfer can be inhibited by blocking relaxase activity within recipient cells with intrabodies. Mol Microbiol 63:404–416. doi: 10.1111/j.1365-2958.2006.05523.x. [DOI] [PubMed] [Google Scholar]
- 59.Jurado P, Fernández LA, De Lorenzo V. 2006. In vivo drafting of single-chain antibodies for regulatory duty on the sigma54-promoter Pu of the TOL plasmid. Mol Microbiol 60:1218–1227. doi: 10.1111/j.1365-2958.2006.05183.x. [DOI] [PubMed] [Google Scholar]
- 60.Zafra O, Fraile S, Gutiérrez C, Haro A, Páez-Espino AD, Jiménez JI, de Lorenzo V. 2011. Monitoring biodegradative enzymes with nanobodies raised in Camelus dromedarius with mixtures of catabolic proteins. Environ Microbiol 13:960–974. doi: 10.1111/j.1462-2920.2010.02401.x. [DOI] [PubMed] [Google Scholar]
- 61.Avalos JL, Fink GR, Stephanopoulos G. 2013. Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols. Nat Biotechnol 31:335–341. doi: 10.1038/nbt.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.DeLoache WC, Dueber JE. 2013. Compartmentalizing metabolic pathways in organelles. Nat Biotechnol 31:320–321. doi: 10.1038/nbt.2549. [DOI] [PubMed] [Google Scholar]
- 63.Heyen U, Schüler D. 2003. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermenter. Appl Microbiol Biotechnol 61:536–544. doi: 10.1007/s00253-002-1219-x. [DOI] [PubMed] [Google Scholar]
- 64.Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- 65.Schultheiss D, Kube M, Schüler D. 2004. Inactivation of the flagellin gene flaA in Magnetospirillum gryphiswaldense results in nonmagnetotactic mutants lacking flagellar filaments. Appl Environ Microbiol 70:3624–3631. doi: 10.1128/AEM.70.6.3624-3631.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heckman KL, Pease LR. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- 67.Raschdorf O, Plitzko JM, Schüler D, Müller FD. 2014. A tailored galK counterselection system for efficient markerless gene deletion and chromosomal tagging in Magnetospirillum gryphiswaldense. Appl Environ Microbiol 80:4323–4330. doi: 10.1128/AEM.00588-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schüler D, Rainer U, Bäuerlein E. 1995. A simple light scattering method to assay magnetism in Magnetospirillum gryphiswaldense. FEMS Microbiol Lett 132:139–145. doi: 10.1111/j.1574-6968.1995.tb07823.x. [DOI] [Google Scholar]
- 69.Laemmli UK. 1970. Cleavage of structural proteins during assembly of head of bacteriophage-T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 70.Popp F, Armitage JP, Schüler D. 2014. Polarity of bacterial magnetotaxis is controlled by aerotaxis through a common sensory pathway. Nat Commun 5:5398. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CheW1-EGFP localization in a dividing cell. Representative differential interference contrast (DIC) and fluorescence micrographs of a cell chromosomally expressing CheW1-EGFP close to cell division. CheW1-EGFP localizes distinctly at the cell poles and forms two new foci at the midcell at the expected position of septum formation. Scale bar, 2 µm. Download
Schematics (a) and fluorescence micrograph of M. gryphiswaldense ΔmamM mutant cells expressing CheW1-EGFP alone (b) or in combination with MamC-2×GBP (c) and wild-type cells expressing chromosomal CheW1-EGFP and cytoplasmic GBP (d) or CheW1-EGFP and MamC-mCherry-GBP (e and f). Scale bar, 2 µm. Download
Western blot of M. gryphiswaldense cells expressing CheW1-EGFP (1), CheW1-EGFP MamC-mCherry-GBP (55.43 kDa) (2), and CheW1-EGFP MamC-2×GBP (41.07 kDa); protein bands are indicated by black arrowheads (3). The native, unfused MamC (12.35 kDa) is present in all strains, indicated by the white arrowhead. Additional bands are visible for all strains expressing MamC fusions, proving stable expression. MamC was detected using rabbit anti-MamC IgG as the primary antibody and goat anti-rabbit IgG alkaline phosphatase antibodies as the secondary antibody. A PageRuler prestained protein ladder from Fermentas was used as a standard. Download
Fluorescence distribution in fluorescence micrographs of M. gryphiswaldense cells expressing chromosomal CheW1-EGFP MamC-2×GBP. Scale bar, 2 µm. Download
TEM micrographs of the M. gryphiswaldense ΔcheW1 strain (a) and cells expressing CheW1-EGFP and cytoplasmic MagGBP (b) or CheW1-EGFP and MamC-mCherry-GBP (c). Scale bar, 1 µm. Download
TEM micrographs of M. gryphiswaldense cells expressing CheW1-EGFP and MamC-2×GBP. Scale bar, 2 µm. Download
Swim halos of M. gryphiswaldense ΔcheOp1 cells and wild-type cells coexpressing either CheW1-EGFP and MagGBPcyt or CheW1-EGFP and MamC-mCherry-GBP. Download
Plasmids used in this study.
Strains used in this study.