Abstract
To determine epidermal growth factor receptor (EGFR) mutation in advanced non-small cell lung cancer (NSCLC) patients and compare the detection efficiency between different sample resources, both high resolution melting (HRM) analysis and direct sequencing method were used to analyze 36 pleural effusion samples and 22 matched biopsy tumor tissues collected from NSCLC patients. For each pleural effusion sample, the supernatant and the cell pellets were examined separately. Among all the 36 cases of pleural effusion samples, 18 mutations of EGFR were found in cell-free supernatant while 13 mutations were found in the cell pellets as detected by HRM analysis. In the 22 matched samples, 13 cases of EGFR mutations were identified in paraffin-embedded biopsy tissue samples, 12 cases in the cell-free supernatant and 9 cases in the cell pellets of pleural effusion. EGFR mutations in 15 cases out of the total 36 pleural effusion samples detected by direct sequencing were also identified by HRM analysis, giving 100% efficiency for HRM method. The results established the important role of HRM as a reliable and efficient method to determine EGFR mutation status and indicated the feasibility of using pleural effusion in replacement of biopsy tissues in particular clinical cases. Furthermore, the cell-free supernatant of pleural effusion might be a better resource for mutation detection than cell pellets.
Keywords: EGFR, NSCLC, pleural effusion, supernatant, HRM
Introduction
Lung cancer is the main cause of cancer-related death all over the world. Non-small cell lung cancer (NSCLC) is the most common form of lung cancer and accounts for about 80% of lung cancer [1,2]. The traditional first-line treatment of advanced NSCLC often involves operative treatment and platinum-based combination chemotherapy [2]. However, due to the lack of overt symptoms, approximately 60-85% of patients are diagnosed in advanced lung cancer period when operative treatment is no more viable and only combination chemotherapy can be applied to inhibit tumor growth, under which circumstance, conventional chemotherapy normally fails to realize long-term therapeutic effect.
In recent years, the epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKI) are widely used in the therapy of NSCLC patients [3-5]. Treatment with these TKIs seems to be especially effective in suspending the development of NSCLC and prolonging life of advanced NSCLC patients [6,7]. However, the curative effect of EGFR-TKI is largely associated with the mutation spectrum and status of epidermal growth factor receptor (EGFR) [6]. Point mutation of exon 18 and exon 21 and deletions of exon 19 in EGFR gene are known to be sensitive for TKIs therapy. About 90% of these mutations are deletions of exon 19 and point mutations of exon 21. Thus, detection of EGFR mutation has become an important event in screening and predicting whether the patient will be benefit from EGFR-TKI targeted therapy or not.
EGFR mutation spectrum of early NSCLC patients can normally be evaluated using samples of surgical removed cancer tissue. However, advanced NSCLC patients who have missed the appropriate time of operative treatment may lose the opportunity to receive drug sensitivity test attributing to unobtainable tumor tissue. Thus the availability of noninvasive diagnostic specimens is of great importance. Pleural effusion is a convenient clinical sample with important clinical diagnostic significance. It may be an alternative source supplying useful information about the mutation status of the EGFR gene. If EGFR gene mutation determination can be achieved with more attainable pleural effusion samples, then targeted drug therapy will be possible for advanced NSCLC patients, which will contribute to vital clinical and practical value [8].
Up to now, different methods have been recruited to detect EGFR mutation and their suitability for EGFR mutation analysis has also been more and more valued [2]. Direct sequencing is a predominant criterion since it is an economical method and can successfully detect all mutations [9,10]. However, direct sequencing is time-consuming and low-sensitivity, limiting its widely application. Hence some faster and more sensitive testing methods are needed. The high resolution melting (HRM) technology has become a hot-spot in the field of life science in recent years [11]. HRM analysis is an attractive screening method based on the physical property of nucleic acid, which adopts saturable dye to monitor the variation of nucleic acid melting curve. New instruments combined with DNA intercalating dyes that can be used at saturating concentrations allow the discrimination of sequence changes in PCR amplicons without manual handling of PCR products [12], making it an ideal candidate to detect DNA sequence changes with advantages of low cost, high throughput, high sensitivity, high specificity and convenience [13].
In this study, we investigate the concordance of EGFR mutations in pleural effusion including cell-free pleural fluid and cellular pellets, and tumor tissue samples from biopsy of the same patients in order to verify the application of pleural effusion in EGFR mutation detection. Both HRM analysis and direct sequencing method were applied and their efficiency is also compared.
Materials and methods
Patients and tumor samples
This study was approved by the Institutional Review Board of Nanfang Hospital, Southern Medical University, and written informed consent was obtained. A total number of 36 cases of malignant pleural effusion and 22 cases of matched tumor tissue samples obtained by thoracoscopic lung biopsy were recruited. The diagnosis of NSCLC was based on cytological or histological findings. The pathological diagnosis was adenocarcinoma in 35 patients and large cell carcinoma in the other patients. There are 16 males and 20 females altogether. The age range was from 31 to 82 ages (median 61.5 years). All individuals in this manuscript have been given written informed consent to publish these case details.
DNA extraction
The pleural effusion samples were centrifugated, then cell-free supernatant and cell pellets were collected respectively. Genomic DNA was extracted by the use of QIAamp DNA Midi Kit according to the manufacturer’s protocols. The tumor tissue samples were embedded by paraffin, and then genomic DNA was extracted by the used of QIAamp DNA FFPE Tissue Kit according to the manufacturer’s protocols.
PCR amplification and HRM for EGFR mutation detection
HRM analysis was carried out by using Human EGFR Gene Mutation Test Kit (HNME-01, Helixgen (Guangzhou) Co. Ltd.) on a LightCyclerTM 480 PCR (Roche Diagnostics) according to the manufacturer’s protocol. The reaction mixture consists of 20 ng of genomic DNA, 300 nM of each primer, 0.5 mM MgCl2 and 1× Master Mix containing LC Green® Plus+ Melting Dye (Biofire Diagnostics) with PCR-grade water adjusted to a final volume of 10 μl. Sample loading was conducted strictly following the manufacturer’s instructions.
The same PCR cycling were applied to all tested exons with an initiate temperature of 95°C for 10 min to activate enzyme and denature template DNA, followed by 50 cycles of 95°C for 20 s, 65°C for 20 s with an initial 10 cycles of touchdown (1°C/cycle) and 72°C for 20 s. The melting step started with a temperature of 95°C for 30 s, which declined to 40°C for 30 s, 75°C for 1 s, and risen up to 95°C again for fluorescence signal captured 20 times per 1°C resulting on a ramp rate of 0.2°C/s. Nucleotide variation was detected based on HRM curve acquisition using the LightCyclerTM 480 Gene Scanning Software (version 1.5). All PCR samples were plotted according to their melting profiles. The normalized graph shows the degree of reduction in fluorescence over a temperature range (75°C to 95°C). Under the difference graph, melting profiles of the samples were compared to that of significant deviations from the horizontal line; those with aberrant melting curves implying the presence of mutation were all recorded as HRM positive. All PCR reactions were performed in duplicate.
The obtained PCR products were purified using QIAquick PCR purification kit and directly sequenced by BGI-Shenzhen to further confirm the genetic variants as detected in HRM analysis.
Statistical analysis
Chi-square test (SPSS Statistics 20.0) was performed to compare the detection efficiency of EGFR mutation between pleural effusion and tumor tissue samples. A P value < 0.05 was considered statistically different. In addition, an inter-rater reliability analysis using the Kappa statistic was performed to determine consistency between the pleural effusion and biopsy samples.
Results
Detection of EGFR mutations by HRM analysis
EGFR mutation status was determined in all the collected 36 pleural effusion samples by HRM analysis. About 18 (18/36, 50%) mutations were detected in the cell-free supernatant and 13 (13/36, 36.1%) mutations in cell pellets (Table 1). EGFR mutations in exon 19 were the most common (13/36, 36.1%), followed by those in exon 21 (5/36; 13.9 %) and exon 20 (1/36; 2.8%). The different plots and sequencing traces of wild type for EGFR exon 19 and exon 21 are shown in Figure 1. No point mutation of exon 18 was detected in this study. The total mutation rate of exon 19 and exon 21 was 50% (18/36). There were discrepancies in the detected mutations between pleural effusion cell pellets and cell-free supernatant. 13 cases of mutation were detected in both cell-free supernatant and cell pellets, while 5 cases of mutation were detected only in cell-free supernatant (Table 1; Figure 2A). Thus, cell-free supernatant is more suitable than cell pellets in EGFR mutation assessment.
Table 1.
Summary of EGFR mutations detected by HRM and sequencing from the 36 NSCLC samples
| Sample | Sex | Age (years) | Histology | EGFR mutation | |||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| HRM | Sequencing | ||||||||
|
| |||||||||
| supernatant | cell pellets | Biopsy tissue | supernatant | cell pellets | Biopsy tissue | ||||
| Case 1 | F | 31 | ADC | 19+ | WT | NA | WT | WT | NA |
| Case 2 | M | 73 | ADC | 19+ | 19+ | NA | 2235_2249del15 | NA | |
| Case 3 | M | 72 | ADC | 21+ | WT | NA | L858R | WT | NA |
| Case 4 | M | 74 | ADC | 19+ | 19+ | NA | 2240_2257del18 | NA | |
| Case 5 | F | 47 | ADC | 19+ | 19+ | NA | 2239_2253del15 | NA | |
| Case 6 | M | 79 | ADC | WT | WT | NA | WT | WT | NA |
| Case 7 | F | 52 | ADC | 19+ | WT | 19+ | 2235_2249del15 | WT | 2235_2249del15 |
| Case 8 | M | 59 | ADC | 20+ | 20+ | 20+ | Ins 773 (DNP) | ||
| Case 9 | F | 48 | ADC | 21+ | 21+ | 21+ | L858R | ||
| Case 10 | F | 36 | LCC | WT | WT | WT | WT | WT | WT |
| Case 11 | M | 70 | ADC | 19+ | 19+ | 19+ | WT | 2239_2248 > C (complex) | |
| Case 12 | F | 56 | ADC | 21+ | WT | 21+ | WT | WT | L858R |
| Case 13 | F | 68 | ADC | 19+ | WT | 19+ | WT | WT | 2235_2249del15 |
| Case 14 | F | 56 | ADC | WT | WT | NA | WT | WT | NA |
| Case 15 | M | 49 | ADC | WT | WT | NA | WT | WT | NA |
| Case 16 | F | 80 | ADC | 21+ | 21+ | NA | L858R | NA | |
| Case 17 | F | 61 | ADC | WT | WT | NA | WT | WT | NA |
| Case 18 | F | 66 | ADC | WT | WT | 21+ | WT | WT | L858R |
| Case 19 | F | 57 | ADC | WT | WT | NA | WT | WT | NA |
| Case 20 | F | 77 | ADC | WT | WT | NA | WT | WT | NA |
| Case 21 | M | 64 | ADC | WT | WT | WT | WT | WT | WT |
| Case 22 | F | 81 | ADC | WT | WT | WT | WT | WT | WT |
| Case 23 | M | 82 | ADC | WT | WT | WT | WT | WT | WT |
| Case 24 | F | 58 | ADC | WT | WT | WT | WT | WT | WT |
| Case 25 | M | 73 | ADC | WT | WT | NA | WT | WT | NA |
| Case 26 | M | 73 | ADC | WT | WT | WT | WT | WT | WT |
| Case 27 | F | 59 | ADC | 19+ | 19+ | 19+ | 2235_2249del15 | ||
| Case 28 | M | 42 | ADC | WT | WT | NA | WT | WT | NA |
| Case 29 | F | 47 | ADC | WT | WT | WT | WT | WT | WT |
| Case 30 | F | 49 | ADC | 19+ | 19+ | 19+ | 2239_2256del18 | ||
| Case 31 | M | 56 | ADC | 19+ | 19+ | 19+ | 2236_2250del15 | ||
| Case 32 | F | 49 | ADC | 19+ | 19+ | 19+ | 2240_2257del18 | ||
| Case 33 | M | 62 | ADC | WT | WT | WT | WT | WT | WT |
| Case 34 | M | 81 | ADC | WT | WT | WT | WT | WT | WT |
| Case 35 | F | 68 | ADC | 19+ | 19+ | 19+ | 2235_2249del15 | ||
| Case 36 | M | 58 | ADC | 19+ | 19+ | 19+ | 2240_2254del15/2239_2253del15 | ||
F, female; M, male; ADC, adenocarcinoma; LCC, large cell carcinoma; WT, wild type; NA, data not available.
Figure 1.
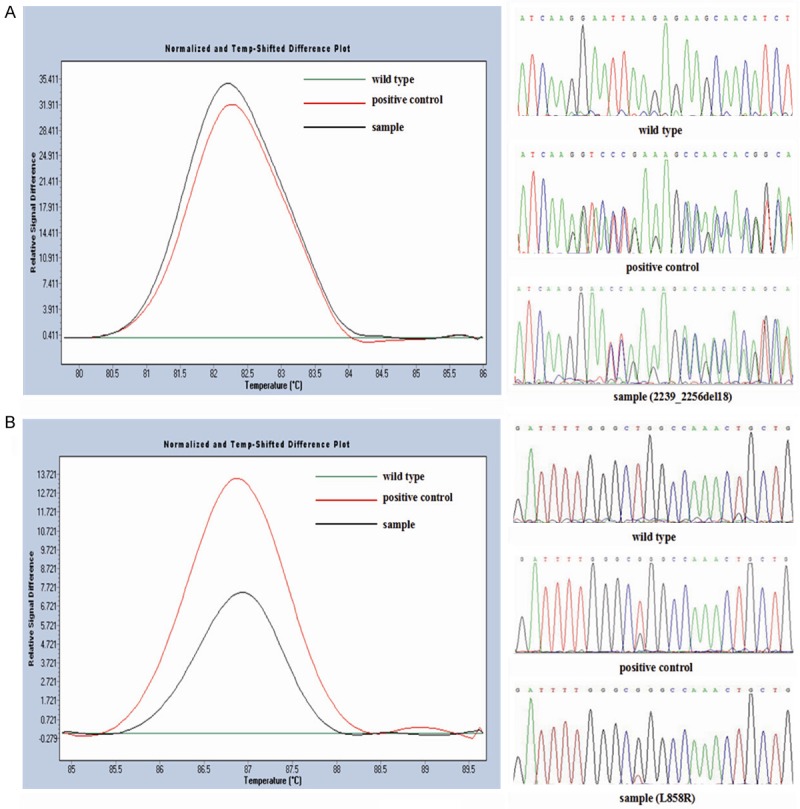
EGFR mutation status detected by HRM analysis and sequencing. The different plots of (A) exon 19 and (B) exon 21 show three different melting profiles corresponding to mutational samples in black, positive control in red and wild type control in green. The results were further confirmed by sequencing.
Figure 2.
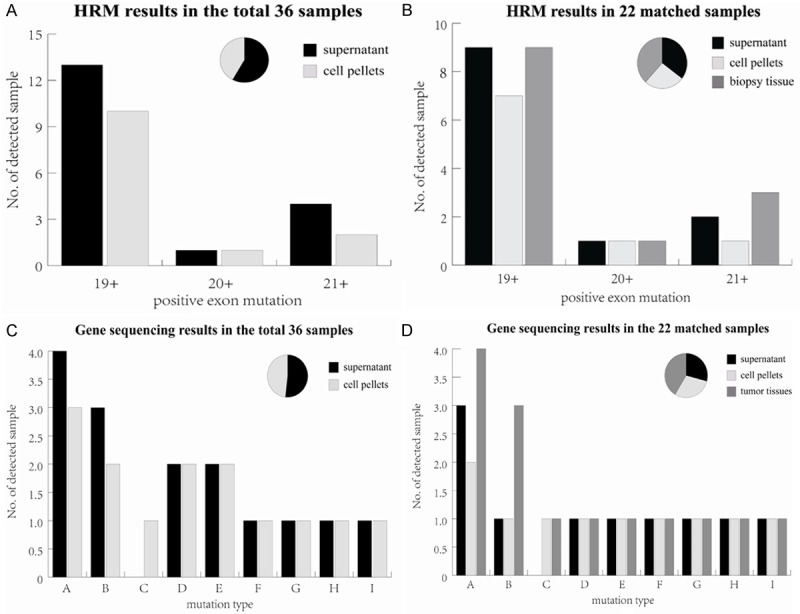
Comparison of EGFR mutation status among different sample resources detected by HRM analysis and sequencing. A. HRM results from the supernatant and cell pellets of pleural effusion in 36 samples; B. HRM results from the supernatant, cell pellets of pleural effusion and tumor tissues in the paired 22 samples; C. Sequencing results from the supernatant and cell pellets of pleural effusions in 36 samples; D. Sequencing results from the supernatant, cell pellets and tumor tissues in the paired 22 samples. The vertical coordinate shows the number of positive mutation samples detected in different EGFR exons or mutation types listed on the horizontal coordinate (exon 18 was not included since no mutation was detected). Overall, 12 cases were detected in both pleural effusion and tumor tissues using HRM method, 1 case of mutation was only detected in tumor tissue samples. Different efficiency degree was revealed in mutation type A, B and C for the three different sample sources. The pie chart displays the total ratio of positive mutation samples detected in all exons or mutation types using different sample sources. Mutations types represented by each capital letter are: A. 2235_2249del15; B. L858R; C. 2239_2248>C (complex); D. 2240_2257del18; E. 2239_2253del15; F. 2240_2254del15; G. 2239_2256del18; H. 2236_2250del15; I. Ins773 (DNP).
In the 22 tumor tissue samples, 13 mutations was detected by HRM, including 9 cases of exon 19 deletion, 3 cases of exon 21 point mutation and 1 case of exon 20 mutation. Among these mutations, 9 mutations were detected in cell pellets and 3 more were detected in the supernatant of pleural effusion, added up to a total of 12 positive mutations detected with pleural effusion. An extra case of mutation was only detected in tumor tissue samples (Table 1; Figure 2B). The consistency of hydrothorax samples and biopsy specimens subjects to 95.5% (21/22) according to HRM results.
Detection of EGFR mutations by sequencing
The PCR products of HRM were all purified and sequenced (Figure 1). EGFR mutations were found in 15 of the 36 pleural effusions. Mutation types encompass deletion mutation in exon 19, L858R in exon 21 and Ins773 (DNP) in exon 20. No missense mutation in exon 18 was found. There are two kinds of deletion mutation in case 36. In total, 15 kinds of mutation were detected in the supernatant of pleural effusion, 14 of which were also detected in cell pellets (Figure 2C). No sample was found to have EGFR mutations by direct sequencing alone (Table 1). Among all these mutations, 2235_2249del15 was observed in 4 cases of tumor tissues, 3 cases of the supernatant of pleural effusion, and only 2 cases of the cell pellets. Mutation L858R was observed in 3 cases of tumor tissues and only 1 case of pleural effusion (Figure 2D). In the 22 cases of paired samples, 3 cases of mutation detected in tumor tissue samples were not successfully detected in pleural effusion, indicating a consistency of 86.4% (19/22). The efficiency of hydrothorax sample resulted in 78.6% (11/14) for both the supernatant and cell pellets.
Comparison of detection efficiency of EGFR mutation status between biopsy and cytological samples
Statistical analysis showed that the kappa values reflecting detection efficiency of EGFR mutation between tumor tissue samples and hydrothorax samples, supernatants of pleural effusion and cell pellets were 0.908 and 0.722 (P < 0.05; Tables 2, 3), respectively. No significant difference was detected between tumor tissue samples and pleural effusion (P > 0.05). The high consistency between these two different sample sources indicates the effectiveness of using cytological samples as a potential substitute of biopsy specimen.
Table 2.
Comparison of detection efficiency of EGFR mutation between 22 pleural effusion samples and matched biopsy tumor tissues by Kappa statistic analysis
| Pleural effusion | ||||
|---|---|---|---|---|
|
| ||||
| + | - | Total | ||
| Tumor tissue | + | 12 | 1 | 13 |
| - | 0 | 9 | 9 | |
| Total | 12 | 10 | 22 | |
+: EGFR positive mutation; -: EGFR negative mutation.
Table 3.
Comparison of detection efficiency of EGFR mutation between the supernatant and cell pellets of pleural effusion samples by Kappa statistic analysis
| Supernatant | ||||
|---|---|---|---|---|
|
| ||||
| + | - | Total | ||
| Cell pellets | + | 13 | 0 | 13 |
| - | 5 | 18 | 23 | |
| Total | 18 | 18 | 36 | |
+: EGFR positive mutation; -: EGFR negative mutation.
Discussion
Nowadays, lung cancer ranks the highest morbidity and mortality among different malignant tumor types and is responsible for approximately 1.38 million deaths each year worldwide [14]. EGFR mutation analysis in NSCLC is a pivotal process during clinic treatment, which can be utilized to predict the patient’s response to EGFR-TKIs [15-17]. Previous studies have proved the therapeutic effect of EGFR-TKI on patients carrying positive EGFR mutations [6,10,18,19]. Therefore, examination of the mutation status of EGFR can provide crucial suggestions on which treatment protocol might be chosen and which patient will be benefit from EGFR targeted therapy along with the prognostic evaluation afterwards [20,21].
However, advanced NSCLC patients heretofore must take biopsy or needle core biopsy to get samples to obtain unequivocal classification diagnosis and genetic detection for targeted drug screening. Concurrently, although paraffin embedded tumor tissue is still the gold-standard sample for EGFR mutation detection and results from molecular analyses using the traditional tumor tissue samples were validated by clinical outcomes [22], it has some restrictions such as inadequate tissue acquisition and non-ideal tissue positions [23]. To overcome the limitation of sample collection in EGFR targeted therapy, numerous studies have employed cytological samples to assess gene mutation, which is in the ascendant currently and has gained favorable clinical effects. In this study, we have selected an atraumatic tumor cell type for patients to whom only adjuvant chemotherapy is available instead of conventional operative treatment, namely, deciduous hydrothorax tumor cell as DNA samples for clinical EGFR mutation detection to conduct targeted drug screening. These cytological samples were added on the basis of tumor tissues as supplement for mutation assessment. Our results show a coherence of 95.5% (21/22) between mutation analyses of 22 paired biopsies and hydrothorax samples from 22 patients. The interrater reliability was found to be statistically significant with a value of Kappa =0.908 (P < 0.05) which indicates fair agreement between these two materials [24]. It offers a kind of tantalizing possibility for advanced NSCLC patients to get the appropriate treatment inferred from EGFR mutation conditions. Now that fresh tissue is not easily accessible for advanced NSCLC patients, cytological samples can serve as a supplement to biopsy specimen as have been suggested by other studies [25-31]. Furthermore, sensitivity methods were also applied in their studies [25-31].
In our study, there was one case of mutation that detected only in tumor tissues but not in cytological sample, showing an efficiency of 92.3% (12/13) for hydrothorax samples. Several explanations might be responsible for this scenario. Firstly, even though HRM analysis was proved to be a suitable methodology to test samples with a low level of DNA content [12], the tumor cells collected in hydrothorax samples were normally far less than surgical samples. Secondly, since malignant mutations do not occur in every single tumor cell, we cannot rule out the possibility of omitting some malignant tumor cells during the acquisition of pleural effusion. In addition, a relative small region or focal distribution of pathological change may lead to failed sampling in a non-optimal puncture site as well. These factors can largely reduced the DNA quantity in pleural fluid to a degree even lower than the resolution capability of HRM analysis. Finally, even though hydrothorax exfoliative cytologic examination is an easily accessible source of specimen with convenient manipulation and comparatively high sensitivity, the veracity of hydrothorax samples has always been labile in different studies. The positive mutation rate detected in pleural fluid ranges from 23% to 70% as reported before [32]. Based on our analyses, the positive mutation rate detected was 92.3% (12/13) by HRM analysis and 78.6% (11/14) by gene sequencing. From the above, we believe that the pleural effusion samples is an ideal substitute of tumor tissues which can provide pivotal evidence for malignance diagnosis under special circumstances, yet commonly, surgical tumor tissue should still be the main sample source. A combined test incorporating cytological samples with biopsy tissue samples can be developed to guarantee the accuracy of EGFR mutation detection.
It is also noteworthy that the positive mutation rate detected from cell-free supernatant of pleural effusions was higher than that detected from cell pellets using either HRM analysis or direct sequencing method in our study. This is probably correlated with a higher content of dissolved tumor DNA in the supernatant. Under high speed of centrifugation, the tumor cells were damaged and DNA molecules were released from the nucleus, making up the dominant components of the supernatant. Even though the pleural effusion often has low tumor cell content than fresh tissues, we can more or less offset this defect by extracting the supernatant part for detection as far as possible. The conformity extrapolates to 83.3% (31/36) between the top phase and the sediment cells in pleural effusions based on HRM results. Our results are in accord with previous study reported by Liu et al [33], suggesting the supernatants of pleural effusion could better reflect the real status of EGFR mutation. Thus, it may provide a more suitable alternative of biopsy tumor tissues than cell pellets.
In the current study, both the direct sequencing and HRM method were recruited to detect EGFR mutations in 36 pleural effusions and the matched 22 cases of tumor tissues. The accuracy of the study was also determined. In the paraffin-embedded tissues, the results of HRM analysis and direct sequencing achieved 100% congruence. While in the pleural effusion samples, HRM method is superior to sequencing method in detection rate with 3 uniquely detected mutations, giving a congruence of 91.7% (33/36). More specifically, these three mutations all came from the supernatant part of pleural effusions with probably higher DNA content than the cell pellets. This evident variance reflected the different efficiency of the two methods and proved that gene sequencing does not support investigations of most cytology samples with low and insufficient tumor cell content. Generally, a threshold of 20% mutant tumor cells was required to be detected by gene sequencing reliably [3,34].
Up to now, high-sensitivity, high-throughput and low-cost technologies are making an impact on genomic research by providing new strategies to fulfill gene mutation determination and scanning, among which the HRM analysis has been specifically noted giving the prominent superiorities of easy operation and wide range of utilization [35,36]. Previous studies have proved that mutant alleles at levels of 1% to 10% can be detected by HRM [3,28]. As a result, it has been widely used in analysis of different cancer- related genes. Gonzalez-Bosquet et al. performed HRM analysis in exons of candidate genes like PIK3CA, ERBB2, KRAS, TP53, EGFR, BRAF, GATA3 and FGFR3 known to harbor established commonly observed mutations [13]. Their results indicating that HRM analysis is a rapid, sensitive and economical method with enormous potential for the detection of DNA sequence changes. Meanwhile, the intrinsic characteristics of HRM endows it with the ability to purify and directly sequence target DNA after analysis without damaging DNA structure, making it a perfect tool of SNP pre-screening before sequencing. However, leaving aside all the advantages, HRM is a robust method that only judges the existence of mutations while no detailed mutation information can be dissected. It cannot completely replace the sequencing method despite of its much higher sensitivity and efficiency. On the contrary, direct sequencing has long been used as a historical standard to detect all known or unknown mutations and provides detailed mutation information. So far, the status of genomic sequence in mutation analysis is still remarkable and positive samples of HRM must be analyzed by direct sequencing to ensure the accuracy. It is expected to be a serious obstacle for HRM method to further develop resulting from its imperfection in clarifying the mutation type and gene sequencing has to be followed up to determine this issue integrally.
In conclusion, our study showed EGFR mutation assessment in pleural effusion samples from NSCLC patients is basically consistent with that in tumor tissue samples obtained by traditional biopsy, indicating that cytological sample is worth to be generalized and adopted clinically. The results also suggested that cell-free supernatant of pleural effusion is a more suitable alternative than cell pellets, but an increased sample size in individual level should be included in further research to confirm this finding. Meanwhile, as a sensitive and reliable method, HRM analysis can be well applied in EGFR mutation detection in somatic cells and mutation screening before sequencing.
Acknowledgements
National Natural Science Foundation of China (Grant No. 30800414) and Pearl River Science & Technology New Star Foundation of Guangzhou City (Grant No. 2012J2200044).
Disclosure of conflict of interest
None.
References
- 1.Zhang X, Zhao Y, Wang M, Yap WS, Chang AY. Detection and comparison of epidermal growth factor receptor mutations in cells and fluid of malignant pleural effusion in non-small cell lung cancer. Lung Cancer. 2008;60:175–182. doi: 10.1016/j.lungcan.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 2.Ellison G, Zhu G, Moulis A, Dearden S, Speake G, McCormack R. EGFR mutation testing in lung cancer: a review of available methods and their use for analysis of tumour tissue and cytology samples. J Clin Pathol. 2013;66:79–89. doi: 10.1136/jclinpath-2012-201194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krypuy M, Newnham GM, Thomas DM, Conron M, Dobrovis A. High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-mall cell lung cancer. BMC Cancer. 2006;6:295. doi: 10.1186/1471-2407-6-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erali M, Voelkerding KV, Wittwer CT. High resolution melting applications for clinical laboratory medicine. Exp Mol Pathol. 2008;85:50–58. doi: 10.1016/j.yexmp.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kramer D, Thunnissen FB, Gallegos-Ruiz MI, Smit EF, Postmus PE, Meijer CJLM, Snijders PJF, Heideman DAM. A fast, sensitive and accurate high resolution melting (HRM) technology-based assay to screen for common K-ras mutations. Cell Oncol. 2009;31:161–167. doi: 10.3233/CLO-2009-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo H, Wan Y, Tian G, Liu Q, Kang Y, Li Y, Yao Z, Lin D. EGFR mutations predict a favorable outcome for malignant leural effusion of lung adenocarcinoma with Tarceva therapy. Oncol Rep. 2012;27:880–890. doi: 10.3892/or.2011.1559. [DOI] [PubMed] [Google Scholar]
- 7.Ikeda T, Nakamura Y, Yamaguchi H, Tomonaga N, Doi S, Nakatomi K, Iida T, Motoshima K, Mizoguchi K, Nagayasu T, Tsukamoto K, Kohno S. Direct Comparison of 3 PCR Methods in Detecting EGFR Mutations in Patients with advanced Non–Small-Cell Lung Cancer. Clin Lung Cancer. 2012;13:369–374. doi: 10.1016/j.cllc.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 8.Yeo CD, Kim JW, Kim KH, Ha JH, Rhee CK, Kim SJ, Kim YK, Park CK, Lee SH, Park MS, Yim HW. Detection and comparison of EGFR mutations in matched tumor tissues, cell blocks, pleural effusions, and sera from patients with NSCLC with malignant pleural effusion, by PNA clamping and direct sequencing. Lung Cancer. 2013;81:207–212. doi: 10.1016/j.lungcan.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 9.Harlé A, Busser B, Rouyer M, Harter V, Genin P, Leroux A, Merlin JL. Comparison of COBAS 4800 KRAS, TaqMan PCR and High Resolution Melting PCR assays for the detection of KRAS somatic mutations in formalin-fixed paraffin embedded colorectal carcinomas. Virchows Arch. 2013;462:329–335. doi: 10.1007/s00428-013-1380-x. [DOI] [PubMed] [Google Scholar]
- 10.Shi Yeen TN, Pathmanathan R, Shiran MS, Ahmad Zaid FA, Cheah YK. Detection of epidermal growth factor receptor mutations in formalin fixed paraffin embedded biopsies in Malaysian non-small cell lung cancer patients. J Biomed Sci. 2013;20:1–7. doi: 10.1186/1423-0127-20-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fassina A, Gazziero A, Zardo D, Corradin M, Aldighieri E, Rossi GP. Detection of EGFR and KRAS mutations on transthoracic needle aspiration of lung nodules by high resolution melting analysis. J Clin Pathol. 2009;62:1096–1102. doi: 10.1136/jcp.2009.067587. [DOI] [PubMed] [Google Scholar]
- 12.Do H, Krypuy M, Mitchell PL, Fox SB, Dobrovic A. High resolution melting analysis for rapid and sensitive EGFR and KRAS mutation detection in formalin fixed paraffin embedded biopsies. BMC Cancer. 2008;8:142. doi: 10.1186/1471-2407-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez-Bosquet J, Calcei J, Wei JS, Garcia-Closas M, Sherman ME, Hewitt S, Vockley J, Lissowska J, Yang HP, Khan J, Chanock S. Detection of Somatic Mutations by High-Resolution DNA Melting (HRM) Analysis in Multiple Cancers. PLoS One. 2011;6:e14522. doi: 10.1371/journal.pone.0014522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 15.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 16.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 17.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H. EGFR receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. PNAS. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller VA, Kris MG, Shah N, Patel J, Azzoli C, Gomez J, Krug LM, Pao W, Rizvi N, Pizzo B, Tyson L, Venkatraman E, Ben-Porat L, Memoli N, Zakowski M, Rusch V, Heelan RT. Bronchioloalveolar pathologic subtype and smoking history predict sensitivity to gefitinib in advanced non-small-cell lung cancer. J. Clin. Oncol. 2004;22:1103–1109. doi: 10.1200/JCO.2004.08.158. [DOI] [PubMed] [Google Scholar]
- 19.Ando M, Okamoto I, Yamamoto N, Takeda K, Tamura K, Seto T, Ariyoshi Y, Fukuoka M. Predictive factors for interstitial lung disease, antitumor response, and survival in non-small cell lung cancer patients treated with gefitinib. J. Clin. Oncol. 2006;24:2549–2556. doi: 10.1200/JCO.2005.04.9866. [DOI] [PubMed] [Google Scholar]
- 20.He M, Capelletti M, Nafa K, Yun CH, Arcila ME, Miller VA, Ginsberg MS, Zhao B, Kris MG, Eck MJ, Jänne PA, Ladanyi M, Oxnard GR. EGFR exon 19 insertions: a new family of sensitizing EGFR mutations in lung adenocarcinoma. Clin Cancer Res. 2012;18:1790–1797. doi: 10.1158/1078-0432.CCR-11-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brevet M, Johnson ML, Azzoli CG, Ladanyi M. Detection of EGFR mutations in plasam DNA from lung cancer patients by mass spectrometry genotyping is predictive of tumor EGFR status and response to EGFR inhibitors. Lung cancer. 2011;73:96–102. doi: 10.1016/j.lungcan.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lozano MD, Zulueta JJ, Cheveste JI, Gúrpide A, Seijo LM, Martín-Algarra S, Del Barrio A, Pio R, Idoate MA, Labiano T, Perez-Gracia JL. Assessment of epidermal growth factor receptor and K-ras mutation status in cytological stained smears of non-small cell lung cancer patients: correlation with clinical outcomes. Oncologist. 2011;16:877–885. doi: 10.1634/theoncologist.2010-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simone G, Mangia A, Malfettone A, Rubini V, Siciliano M. Chromogenic in situ hybridization to detect EGFR gene copy number in cell blocks from fine-needle aspirates of non small cell lung carcinomas and lung metastases from colo-rectal cancer. J Exp Clin Cancer Res. 2010;29:125. doi: 10.1186/1756-9966-29-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landis JR, Koch GG. The measurement of observer agreement for Galegorical data. Biometrics. 1997;34:209–212. [PubMed] [Google Scholar]
- 25.da Cunha Santos G, Saieg MA, Geddie W, Leighl N. EGFR gene status in cytological samples of nonsmall cell lung carcinoma: controversies and opportunities. Ann Oncol. 2011;119:80–91. doi: 10.1002/cncy.20150. [DOI] [PubMed] [Google Scholar]
- 26.Goto K, Satouchi M, Ishii G, Nishio K, Hagiwara K, Mitsudomi T, Whiteley J, Donald E, McCormack R, Todo T. An evaluation study of EGFR mutation tests utilized for non-small-cell lung cancer in the diagnostic setting. Ann Oncol. 2012;23:2914–2919. doi: 10.1093/annonc/mds121. [DOI] [PubMed] [Google Scholar]
- 27.Boldrini L, Gisfredi S, Ursino S, Camacci T, Baldini E, Melfi F, Fontanini G. Mutational analysis in cytological specimens of advanced lung adenocarcinoma: A sensitive method for molecular diagnosis. J Thorac Oncol. 2007;2:1086–1090. doi: 10.1097/JTO.0b013e31815ba1fa. [DOI] [PubMed] [Google Scholar]
- 28.Nomoto K, Tsuta K, Takano T, Fukui T, Yokozawa K. Detection of EGFR mutations in archived cytologic specimens of non-small cell lung cancer using high resolution melting analysis. Am J Clin Pathol. 2006;126:608–615. doi: 10.1309/N5PQNGW2QKMX09X7. [DOI] [PubMed] [Google Scholar]
- 29.Smith GD, Chadwick BE, Willmore-Payne C, Bentz JS. Detection of epidermal growth factor receptor gene mutations in cytology specimens from patients with non-small cell lung cancer utilising high-resolution melting amplicon analysis. J Clin Pathol. 2008;61:487–493. doi: 10.1136/jcp.2007.051425. [DOI] [PubMed] [Google Scholar]
- 30.Oshita F, Matsukuma S, Yoshihara M, Sakuma Y, Ohgane N, Kameda Y, Saito H, Yamada K, Tsuchiya E, Miyagi Y. Novel heteroduplex method using small cytology specimens with a remarkably high success rate for analysing EGFR gene mutations with a significant correlation to gefitinib efficacy in non-small-cell lung cancer. Br J Cancer. 2006;95:1070–1075. doi: 10.1038/sj.bjc.6603396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Billah S, Stewart J, Staerkel G, Chen S, Gong Y, Guo M. EGFR and KRAS mutations in lung carcinoma. Cancer Cytopathol. 2011;119:111–117. doi: 10.1002/cncy.20151. [DOI] [PubMed] [Google Scholar]
- 32.Attanoos RL, Gibbs AR. The comparative accuracy of different pleural biopsy techniques in the diagnosis of malignant mesothelioma. Histopathology. 2008;53:340–344. doi: 10.1111/j.1365-2559.2008.03099.x. [DOI] [PubMed] [Google Scholar]
- 33.Liu D, Lu Y, Hu Z, Wu N, Nie X, Xia Y, Han Y, Li Q, Zhu G, Bai C. Malignant Pleural Effusion Supernatants Are Substitutes for Metastatic Pleural Tumor Tissues in EGFR Mutation Test in Patients with Advanced Lung Adenocarcinoma. PLoS One. 2014;9:e89946. doi: 10.1371/journal.pone.0089946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogino S, Kawasaki T, Brahmandam M, Yan L, Cantor M, Namgyal C, Mino-Kenudson M, Lauwers GY, Loda M, Fuchs CS. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7:413–421. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guedes JG, Veiga I, Rocha P, Pinto P, Pinto C, Pinheiro M, Peixoto A, Fragoso M, Raimundo A, Ferreira P, Machado M, Sousa N, Lopes P, Araújo A, Macedo J, Alves F, Coutinho C, Henrique R, Santos LL, Teixeira MR. High resolution melting analysis of KRAS, BRAF and PIK3CA in KRAS exon 2 wild-type metastatic colorectal cancer. BMC Cancer. 2013;13:169. doi: 10.1186/1471-2407-13-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Obul J, Itoga S, Abliz M, Sato K, Ishige T, Utsuno E, Matsushita K, Matsubara H, Nomura F. High-resolution melting analyses for gene scanning of APC, MLH1, MSH2, and MSH6 associated with hereditary colorectal cancer. Genet Test Mol Bioma. 2012;16:406–411. doi: 10.1089/gtmb.2011.0166. [DOI] [PubMed] [Google Scholar]


