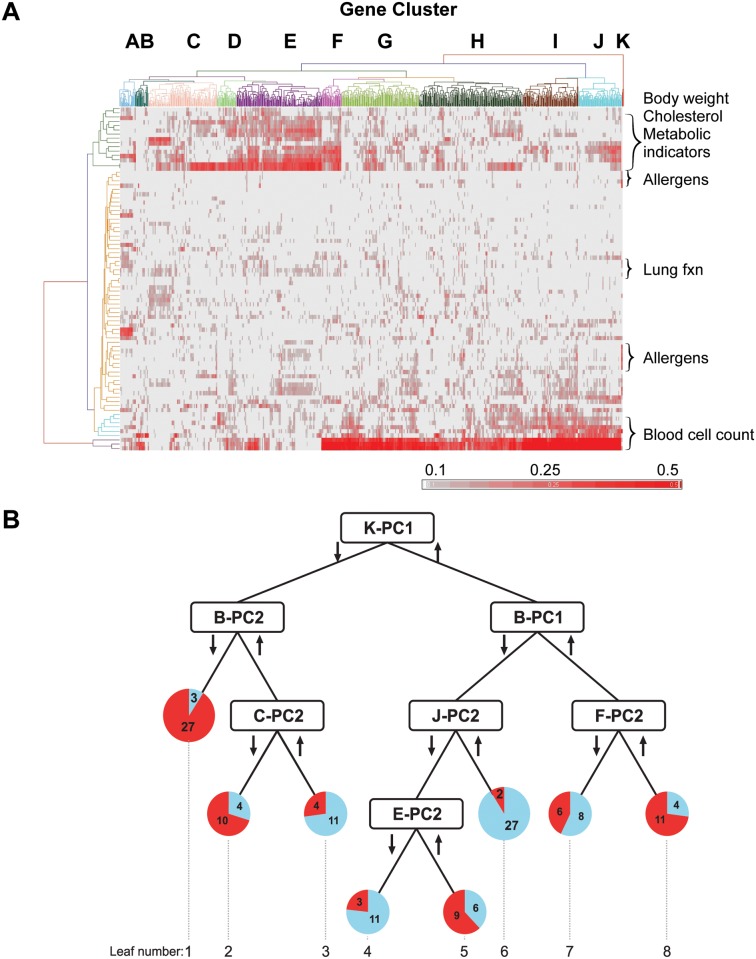
Fig 1. Data integration and reduction to build decision tree.
Adapted from [23]. (A) Heat map shows absolute value of the Pearson correlations between 901 genes (X axis) and 81 clinical biomarkers (Y axis) for the 192 study subjects. Hierarchical clustering yielded 11 gene clusters labeled A-K with the corresponding gene lists provided in S5 Table. The clinical biomarkers are listed in S6 Table along with their dendrogram-clustered groupings. (B) Decision tree shows partitioning of the 146 subjects with unambiguous asthma status into mechanistically distinct asthmatic and non-asthmatic leaves based on metagenes developed by dimension reduction of the gene clusters using principal component analysis. The metagenes are labeled by gene cluster and principal component (e.g., K-PC1 represents gene cluster K, principal component 1). Arrows represent whether subjects were above or below the decision tree’s entropy-based cutpoint. Pie charts for each leaf show the number of asthmatics (red) and non-asthmatics (blue). Geometric means of selected clinical biomarkers per leaf are provided in S3 and S4 Tables.