Figure 1. Residue N242 is required for LegC7 toxicity in yeast.
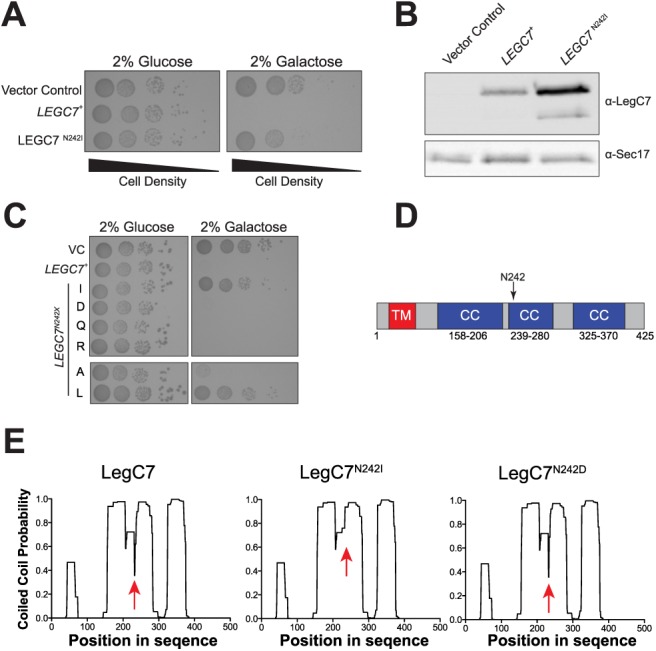
(A) BY4742 yeast strains harboring the galactose-inducible control plasmid pYES2/NT C, pVJS52 (LEGC7 +), or pVJS54 (LEGC7 N242I) were spotted onto CSM-uracil medium supplemented with either 2% glucose or 2% galactose with 10-fold serial dilutions from a starting culture of OD600 = 1.0. Plates were incubated for 72 h at 30°C. (B) Strains from (A) were grown in for 24 h in CSM-uracil supplemented with 2% glucose at 30°C, washed in ddH2O, suspended in fresh CSM-uracil/2% galactose, and incubated at 30°C for 16 h. Equal fractions of each strain were harvested, total protein was extracted [55], and 30μl from each sample was separated by SDS-PAGE. Samples were immunoblotted for LegC7 (rabbit 1:5000) or Sec17p (Rabbit, 1:1000) [56] (loading control). (C) The LEGC7 + plasmid, pVJS52, was mutagenized via site-directed mutagenesis (Materials and Methods), transformed into BY4742, and spotted onto CSM-Ura medium containing either 2% glucose or 2% galactose in 10-fold serial dilutions. (D) Diagram of the predicted LegC7 protein structure indicating transmembrane domain (TM, red) and three predicted coiled coil domains (CC, blue). Transmembrane prediction was calculated with TMHMM Server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/,) and coiled coil predictions were calculated with COILS (http://toolkit.tuebingen.mpg.de/pcoils) with a window size of 21, weighting, and an iterated matrix. (E) Coiled coil probability prediction of LegC7 containing either N, I, or D at position 242 were run as in (D). Probabilities at each position were plotted and the predicted disordered region between predicted coiled coil regions 1 and 2 is marked (red arrow).
