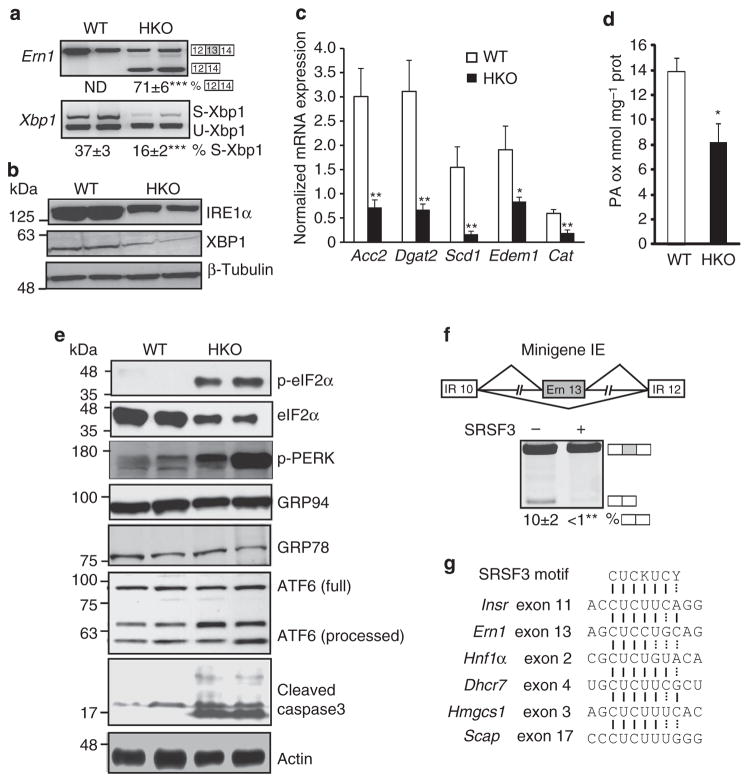
Figure 6. Loss of SRSF3 dysregulates the UPR.
(a) Analysis of Xbp1 and Ern1 splicing. A representative gel is shown with two WT and two SRSF3HKO samples and the percentage of exon 13 skipping for Ern1 and spliced Xbp1 (S-Xbp1) is shown below. S-Xbp1 and U-Xbp1 indicate the spliced and unspliced form of Xbp1, respectively. (b) XBP1 and IRE1α protein expression by immunoblotting. Blots were stripped and reblotted for β-tubulin. (c) mRNA expression of XBP1 target genes Acc2, Dgat2, Scd1, Edem1 and Cat in SRSF3HKO and WT mice by qPCR; n = 8. (d) Fatty acid oxidation in liver lysates from SRSF3HKO and WT mice; n = 3–4. (e) eIF2α and PERK phosphorylation, GRP78 and GRP94 expression, and ATF6 and caspase-3 processing and cleavage measured by immunoblotting SRSF3HKO and WT liver lysates. Blots were stripped and reblotted for eIF2α and actin as loading controls. (f) Schematic drawing of a minigene containing INSR exons 10 and 12 (white boxes) and Ern1 exon 13 (grey box). Representative gel shows exon 13 skipping in RNA from NIH3T3 cells transfected with the minigene with or without a SRSF3 expression plasmid (n = 3 expts). (g) Consensus binding motif of SRSF3 is present in exons of Insr, Ern1, Hnf1α, Dhcr7 and Scap and also in the downstream exon 3 of Hmgcs1. Results are means±s.e.m. *P<0.05 and **P<0.01, and ***P<0.001.